What Pediatricians Should Know before Studying Gut Microbiota
Abstract
:1. Introduction
2. Basic Knowledge on Gut Microbes
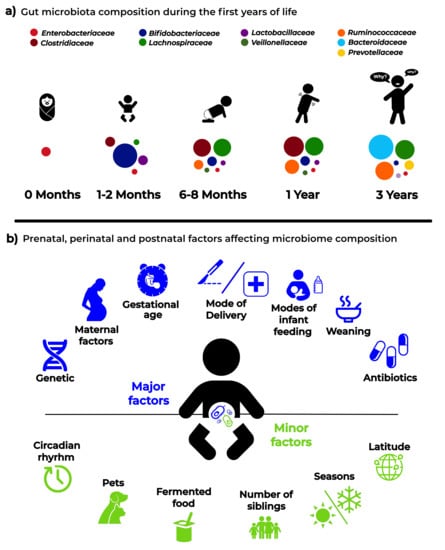
3. The Intestinal Microbiota from Birth Throughout Childhood
4. Issues to be Considered for Studying Microbiome in Clinical Studies
Study Design and Patient Selections
5. Major Pre-, Peri-, and Post-Natal Factors Affecting the Child Gut Microbiota
5.1. Maternal Factors Influencing Infant Microbiota
5.1.1. Changes Related to Vertical Transmission of Maternal Metabolites
5.1.2. Changes Related to Dietary Patterns and Lifestyle
5.2. Genetic Factors
5.3. Mode of Delivery
5.4. Mode of Infant Feeding
5.5. Gestational Age
5.6. Antibiotics
5.7. Weaning
6. Minor Factors Affecting Gut Microbiota
7. Sample Collection
8. Discussion
9. Conclusions and Future Perspectives
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Zmora, N.; Zilberman-Schapira, G.; Suez, J.; Mor, U.; Dori-Bachash, M.; Bashiardes, S.; Kotler, E.; Zur, M.; Regev-Lehavi, D.; Brik, R.B.Z.; et al. Personalized Gut Mucosal Colonization Resistance to Empiric Probiotics Is Associated with Unique Host and Microbiome Features. Cell 2018, 174, 1388–1405. [Google Scholar] [CrossRef] [PubMed]
- Yatsunenko, T.; Rey, F.E.; Manary, M.J.; Trehan, I.; Dominguez-Bello, M.G.; Contreras, M.; Magris, M.; Hidalgo, G.; Baldassano, R.N.; Anokhin, A.P.; et al. Human gut microbiome viewed across age and geography. Nature 2012, 486, 222–227. [Google Scholar] [CrossRef] [PubMed]
- Rinninella, E.; Raoul, P.; Cintoni, M.; Franceschi, F.; Miggiano, G.A.D.; Gasbarrini, A.; Mele, M.C. What is the Healthy Gut Microbiota Composition? A Changing Ecosystem across Age, Environment, Diet, and Diseases. Microorganisms 2019, 7, 14. [Google Scholar] [CrossRef] [PubMed]
- Lozupone, C.A.; Stombaugh, J.I.; Gordon, J.I.; Jansson, J.K.; Knight, R. Diversity, stability and resilience of the human gut microbiota. Nature 2012, 489, 220–230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williams, S.C. The other microbiome. Proc. Natl. Acad. Sci. USA 2013, 110, 2682–2684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomas, T.; Gilbert, J.; Meyer, F. Metagenomics—A guide from sampling to data analysis. Microb. Inform. Exp. 2012, 2, 3. [Google Scholar] [CrossRef] [PubMed]
- Turnbaugh, P.J.; Gordon, J.I. An invitation to the marriage of metagenomics and metabolomics. Cell 2006, 134, 708–713. [Google Scholar] [CrossRef]
- Trivedi, B. Microbiome: The surface brigade. Nature 2012, 492, S60–S61. [Google Scholar] [CrossRef]
- Lagier, J.C.; Dubourg, G.; Million, M.; Cadoret, F.; Bilen, M.; Fenollar, F.; Levasseur, A.; Rolain, J.M.; Fournier, P.E.; Raoult, D. Culturing the human microbiota and culturomics. Nat. Rev. Genet. 2018, 16, 540–550. [Google Scholar] [CrossRef] [Green Version]
- Peterson, J.; Garges, S.; Giovanni, M.; McInnes, P.; Wang, L.; Schloss, J.A.; Bonazzi, V.; McEwen, J.E.; Wetterstrand, K.A.; Deal, C.; et al. The NIH human microbiome project. Genome Res. 2009, 19, 2317–2323. [Google Scholar]
- Costello, E.K.; Lauber, C.L.; Hamady, M.; Fierer, N.; Gordon, J.I.; Knight, R. Bacterial Community Variation in Human Body Habitats Across Space and Time. Science 2009, 326, 1694–1697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qin, J.; Li, R.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; Yamada, T.; et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010, 464, 59–65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kelly, B.J.; Gross, R.; Bittinger, K.; Sherrill-Mix, S.; Lewis, J.D.; Collman, R.G.; Bushman, F.D.; Li, H. Power and sample-size estimation for microbiome studies using pairwise distances and PERMANOVA. Bioinformatics 2015, 31, 2461–2468. [Google Scholar] [CrossRef] [Green Version]
- Arrieta, M.C.; Stiemsma, L.T.; Amenyogbe, N.; Brown, E.M.; Finlay, B. The Intestinal Microbiome in Early Life: Health and Disease. Front. Immunol. 2014, 5, 427. [Google Scholar] [CrossRef] [Green Version]
- Milani, C.; Duranti, S.; Bottacini, F.; Casey, E.; Turroni, F.; Mahony, J.; Belzer, C.; Palacio, S.D.; Montes, S.A.; Mancabelli, L.; et al. The First Microbial Colonizers of the Human Gut: Composition, Activities, and Health Implications of the Infant Gut Microbiota. Microbiol. Mol. Biol. Rev. 2017, 81, e00036-17. [Google Scholar] [CrossRef]
- Willyard, C. Could baby’s first bacteria take root before birth? Nature 2018, 553, 264–266. [Google Scholar] [CrossRef]
- Matamoros, S.; Guen, C.G.L.; Le Vacon, F.; Potel, G.; De La Cochetière, M.F. Development of intestinal microbiota in infants and its impact on health. Trends Microbiol. 2013, 21, 167–173. [Google Scholar] [CrossRef]
- Hill, C.J.; Lynch, D.B.; Murphy, K.; Ulaszewska, M.; Jeffery, I.B.; O’Shea, C.A.; Watkins, C.; Dempsey, E.; Mattivi, F.; Touhy, K.; et al. Evolution of gut microbiota composition from birth to 24 weeks in the INFANTMET Cohort. Microbiome 2017, 5, 4. [Google Scholar] [CrossRef]
- Stewart, C.J.; Ajami, N.J.; O’Brien, J.L.; Hutchinson, D.S.; Smith, D.P.; Wong, M.C.; Ross, M.C.; Lloyd, R.E.; Doddapaneni, H.; Metcalf, G.A.; et al. Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature 2018, 562, 583. [Google Scholar] [CrossRef]
- Falony, G.; Joonssens, M.; Vieine-Silva, S.; Wang, J.; Darzi, Y.; Faust, K.; Kurilshikov, A.; Bonder, M.J.; Valles-Colomer, M.; Vandeputte, P.; et al. Population-level analysis of gut microbiota variation. Science 2016, 352, 560–564. [Google Scholar] [CrossRef] [PubMed]
- De Agüero, M.G.; Ganal-Vonarburg, S.C.; Fuhrer, T.; Rupp, S.; Uchimura, Y.; Li, H.; Steinert, A.; Heikenwalder, M.; Hapfelmeier, S.; Sauer, U.; et al. The maternal microbiota drives early postnatal innate immune development. Science 2016, 351, 1296–1302. [Google Scholar] [CrossRef] [PubMed]
- Rakoff-Nahoum, S. Another reason to thank mum: Gestational effects of microbiota metabolites. Cell Host Microbe 2016, 19, 425–427. [Google Scholar] [CrossRef] [PubMed]
- MacPherson, A.J.; De Agüero, M.G.; Ganal-Vonarburg, S.C. How nutrition and the maternal microbiota shape the neonatal immune system. Nat. Rev. Immunol. 2017, 17, 508–517. [Google Scholar] [CrossRef] [PubMed]
- Kearney, J.F.; Patel, P.; Stefanov, E.K.; King, R.G. Natural Antibody Repertoires: Development and Functional Role in Inhibiting Allergic Airway Disease. Annu. Rev. Immunol. 2015, 33, 475–504. [Google Scholar] [CrossRef] [PubMed]
- Bonder, M.J.; Kurilshikov, A.; Tigchelaar, E.F.; Mujagic, Z.; Imhann, F.; Vila, A.V.; Deelen, P.; Vatanen, T.; Schirmer, M.; Smeekens, S.P.; et al. The effect of host genetics on the gut microbiome. Nat. Genet. 2016, 48, 1407–1412. [Google Scholar] [CrossRef] [PubMed]
- Goodrich, J.K.; Waters, J.L.; Poole, A.C.; Sutter, J.L.; Koren, O.; Blekhman, R.; Beaumont, M.; Van Treuren, W.; Knight, R.; Bell, J.T.; et al. Human genetics shape the gut microbiome. Cell 2014, 159, 789–799. [Google Scholar] [CrossRef] [PubMed]
- Dominguez-Bello, M.G.; Costello, E.K.; Contreras, M.; Magris, M.; Hidalgo, G.; Fierer, N.; Knight, R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. USA 2010, 107, 11971–11975. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fallani, M.; Young, D.; Scott, J.; Norin, E.; Amarri, S.; Adam, R.; Aguilera, M.; Khanna, S.; Gil, A.; Edwards, A.C.; et al. Intestinal Microbiota of 6-week-old Infants Across Europe: Geographic Influence Beyond Delivery Mode, Breast-feeding, and Antibiotics. J. Pediatr. Gastroenterol. Nutr. 2010, 51, 77–84. [Google Scholar] [CrossRef]
- Drago, L.; Toscano, M.; De Grandi, R.; Grossi, E.; Padovani, E.M.; Peroni, D.G. Microbiota network and mathematic microbe mutualism in colostrum and mature milk collected in two different geographic areas: Italy versus Burundi. ISME J. 2017, 11, 875–884. [Google Scholar] [CrossRef]
- Groer, M.W.; Luciano, A.A.; Dishaw, L.J.; Ashmeade, T.L.; Miller, E.; Gilbert, A.J. Development of the preterm infant gut microbiome: A research priority. Microbiome 2014, 2, 38. [Google Scholar] [CrossRef] [PubMed]
- Stewart, C.J.; Embleton, N.D.; Clements, E.; Luna, P.N.; Smith, D.P.; Fofanova, T.Y.; Nelson, A.; Taylor, G.; Orr, C.H.; Petrosino, J.F.; et al. Cesarean or Vaginal Birth Does Not Impact the Longitudinal Development of the Gut Microbiome in a Cohort of Exclusively Preterm Infants. Front. Microbiol. 2017, 8, 1008. [Google Scholar] [CrossRef] [PubMed]
- Brooks, B.; Olm, M.R.; Firek, B.A.; Baker, R.; Thomas, B.C.; Morowitz, M.J.; Banfield, J.F. Strain-resolved analysis of hospital rooms and infants reveals overlap between the human and room microbiome. Nat. Commun. 2017, 8, 1814. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Johani, K.; Gosbell, I.; Jacombs, A.; Almatroudi, A.; Whiteley, G.; Deva, A.; Jensen, S.; Vickery, K. Intensive care unit environmental surfaces are contaminated by multidrug-resistant bacteria in biofilms: Combined results of conventional culture, pyrosequencing, scanning electron microscopy, and confocal laser microscopy. J. Hosp. Infect. 2015, 91, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Wandro, S.; Osborne, S.; Enriquez, C.; Bixby, C.; Arrieta, A.; Whiteson, K. The microbiome and metabolome of preterm onfant stool are personalized and not driven by health outcomes, including necrotizing enterocolitis and late-onset sepsis. Msphere 2018, 3, e00104–e00118. [Google Scholar] [CrossRef] [PubMed]
- Fouhy, F.; Guinane, C.M.; Hussey, S.; Wall, R.; Ryan, C.A.; Dempsey, E.M.; Murphy, B.; Ross, R.P.; Fitzgerald, G.F.; Stanton, C.; et al. High-Throughput Sequencing Reveals the Incomplete, Short-Term Recovery of Infant Gut Microbiota following Parenteral Antibiotic Treatment with Ampicillin and Gentamicin. Antimicrob. Agents Chemother. 2012, 56, 5811–5820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanaka, S.; Kobayashi, T.; Songjinda, P.; Tatejama, A.; Tsubouchi, M.; Kiyohara, C.; Shirakawa, T.; Sonomoto, J.; Nakayama, J. Influence of antibiotic exposure in the early postnatal period in the development of intestinal microbiota. FEMS Immunol. Med. Microbiol. 2009, 56, 80–87. [Google Scholar] [CrossRef]
- Dethlefsen, L.; Relman, D.A. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc. Natl. Acad. Sci. USA 2011, 108, 4554–4561. [Google Scholar] [CrossRef]
- Koenig, J.E.; Spor, A.; Scalfone, N.; Fricker, A.D.; Stombaugh, J.; Knight, R.; Angenent, L.T.; Ley, R.E. Succession of microbial consortia in the developing infant gut microbiome. Proc. Natl. Acad. Sci. USA 2011, 108, S4578–S4585. [Google Scholar] [CrossRef]
- Thaiss, C.A.; Levy, M.; Korem, T.; Dohnalová, L.; Shapiro, H.; Jaitin, D.A.; David, E.; Winter, D.R.; Gury-BenAri, M.; Tatirovsky, E.; et al. Microbiota Diurnal Rhythmicity Programs Host Transcriptome Oscillations. Cell 2016, 167, 1495–1510. [Google Scholar] [CrossRef]
- Misic, A.M.; Davis, M.F.; Tyldsley, A.S.; Hodkinson, B.P.; Tolomeo, P.; Hu, B.; Nachamkin, I.; Lautenbach, E.; Morris, O.D.; Grice, A.E. The shared microbiota of humans and companion animals as evaluated from Staphylococcus carriage sites. Microbiome 2015, 3, 2. [Google Scholar] [CrossRef] [PubMed]
- Davenport, E.R.; Mizrahi-Man, O.; Michelini, K.; Barreiro, L.B.; Ober, C.; Gilad, Y. Seasonal Variation in Human Gut Microbiome Composition. PLoS ONE 2014, 9, e90731. [Google Scholar] [CrossRef] [PubMed]
- Costea, P.I.; Zeller, G.; Sunagawa, S.; Pelletier, E.; Alberti, A.; Levenez, F.; Tramontano, M.; Driessen, M.; Hercog, R.; Jung, F.E.; et al. Towards standards for human fecal sample processing in metagenomic studies. Nat. Biotechnol. 2017, 35, 1069–1076. [Google Scholar] [CrossRef] [PubMed]
- Jordan, S.; Baker, B.; Dunn, A.; Edwards, S.; Ferranti, E.; Mutic, A.D.; Yang, I.; Rodriguez, J. Maternal–Child Microbiome: Specimen Collection, Storage and Implications for Research and Practice. Nurs. Res. 2017, 66, 175–183. [Google Scholar] [CrossRef] [PubMed]
- Tedjo, D.I.; Jonkers, D.M.A.E.; Savelkoul, P.H.; Masclee, A.A.; Van Best, N.; Pierik, M.J.; Penders, J. The Effect of Sampling and Storage on the Fecal Microbiota Composition in Healthy and Diseased Subjects. PLoS ONE 2015, 10, 0126685. [Google Scholar] [CrossRef] [PubMed]
- Choo, J.M.; Leong, L.E.; Rogers, G.B. Sample storage conditions significantly influence faecal microbiome profiles. Sci. Rep. 2015, 5, 16350. [Google Scholar] [CrossRef]
- Bassis, C.M.; Moore, N.M.; Lolans, K.; Seekatz, A.M.; Weinstein, R.A.; Young, V.B.; Hayden, M.K. CDC Prevention Epicenters Program. Comparison of stool versus rectal swab samples and storage conditions on bacterial community profiles. BMC Microbiol. 2017, 17, 78–85. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Maldonado, J.; Kang, D.W.; Krajmalnik-Brown, R.; Caporaso, J.G. Rapidly Processed Stool Swabs Approximate Stool Microbiota Profiles. mSphere 2019, 4, e00208–e00219. [Google Scholar] [CrossRef]
- Lauder, A.P.; Roche, A.M.; Sherrill-Mix, S.; Bailey, A.; Laughlin, A.L.; Bittinger, K.; Leite, R.; Elovitz, M.A.; Parry, S.; Bushman, F.D. Comparison of placenta samples with contamination controls does not provide evidence for a distinct placenta microbiota. Microbiome 2016, 4, 29. [Google Scholar] [CrossRef]
- Perez-Munoz, M.E.; Arrieta, M.C.; Ramer-Tai, A.E.; Walter, J. A critical assessment of the “sterile womb” and “in utero colonization” hypothesis: Implications for research of the pioneer infant microbiome. Microbiome 2017, 5, 48–67. [Google Scholar] [CrossRef]
- Panelli, S.; Schneider, L.; Comandatore, F.; Bandi, C.; Zuccotti, G.V.; D’Auria, E.; Dauria, E. Is there life in the meconium? A challenging, burning question. Pharmacol. Res. 2018, 137, 148–149. [Google Scholar] [CrossRef] [PubMed]
- Ferri, E.; Genco, F.; Gulminetti, R.; Bandi, C.; Novati, S.; Casiraghi, M.; Sambri, V. Plasma Levels of Bacterial DNA in HIV Infection: The Limits of Quantitative Polymerase Chain Reaction. J. Infect. Dis. 2010, 202, 176–177. [Google Scholar] [CrossRef] [PubMed]
- Zaneveld, J.R.; McMinds, R.; Thurber, R.V. Stress and stability: Applying the Anna Karenina principle to animal microbiomes. Nat. Microbiol. 2017, 2, 17121. [Google Scholar] [CrossRef] [PubMed]


| Stages and Pitfalls | Considerations and Practical Solutions |
|---|---|
| Study question |
|
| Statistically underpowered studies |
|
| Selection of subjects: avoiding heterogeneity of the population |
|
| Confounding factors (lifestyle and clinical factors) |
|
| Timing and frequency of sample collection |
|
| Sample collection and storage |
|
| Experimental Lab procedures |
|
| Sequencing |
|
| Data analysis |
|
| Risk-benefit assessment |
|
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Drago, L.; Panelli, S.; Bandi, C.; Zuccotti, G.; Perini, M.; D’Auria, E. What Pediatricians Should Know before Studying Gut Microbiota. J. Clin. Med. 2019, 8, 1206. https://0-doi-org.brum.beds.ac.uk/10.3390/jcm8081206
Drago L, Panelli S, Bandi C, Zuccotti G, Perini M, D’Auria E. What Pediatricians Should Know before Studying Gut Microbiota. Journal of Clinical Medicine. 2019; 8(8):1206. https://0-doi-org.brum.beds.ac.uk/10.3390/jcm8081206
Chicago/Turabian StyleDrago, Lorenzo, Simona Panelli, Claudio Bandi, Gianvincenzo Zuccotti, Matteo Perini, and Enza D’Auria. 2019. "What Pediatricians Should Know before Studying Gut Microbiota" Journal of Clinical Medicine 8, no. 8: 1206. https://0-doi-org.brum.beds.ac.uk/10.3390/jcm8081206