Molecular Profiling of Malignant Pleural Effusions with Next Generation Sequencing (NGS): Evidence that Supports Its Role in Cancer Management
Abstract
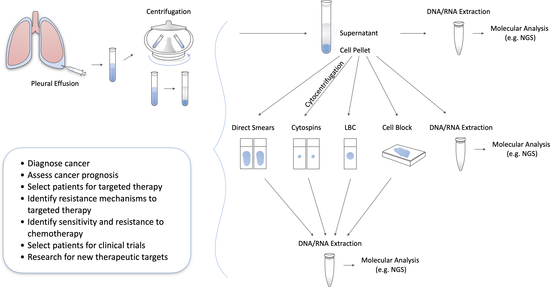
:1. Introduction
2. Pleural Fluid Liquid Biopsy Compared to Other Types of Samples
3. Correlation of Pleural Fluid NGS with Cytomorphologic Findings and Tumor Cellularity
4. The Value of Supernatant-Derived cfDNA
5. Evaluation of Therapeutic Resistance, Response, and Management
6. The Role of NGS in Malignant Pleural Effusions from Different Types of Cancers (Other than Lung)
7. Discussion
8. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Yalcin, N.G.; Choong, C.K.C.; Eizenberg, N. Anatomy and pathophysiology of the pleura and pleural space. Thorac. Surg. Clin. 2013, 23, 1–10. [Google Scholar] [CrossRef]
- Engels, M.; Michael, C.; Dobra, K.; Hjerpe, A.; Fassina, A.; Firat, P. Management of cytological material, pre-analytical procedures and bio-banking in effusion cytopathology. Cytopathology 2019, 30, 31–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Psallidas, I.; Kalomenidis, I.; Porcel, J.M.; Robinson, B.W.; Stathopoulos, G.T. Malignant pleural effusion: From bench to bedside. Eur. Respir. Rev. 2016, 25, 189–198. [Google Scholar] [CrossRef] [PubMed]
- Sundling, K.E.; Cibas, E.S. Ancillary studies in pleural, pericardial, and peritoneal effusion cytology. Cancer Cytopathol. 2018, 126, 590–598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morgensztern, D.; Waqar, S.; Subramanian, J.; Trinkaus, K.; Govindan, R. Prognostic impact of malignant pleural effusion at presentation in patients with metastatic non-small-cell lung cancer. J. Thorac. Oncol. 2012, 7, 1485–1489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bibby, A.C.; Tsim, S.; Kanellakis, N.; Ball, H.; Talbot, D.C.; Blyth, K.G.; Maskell, N.A.; Psallidas, I. Malignant pleural mesothelioma: An update on investigation, diagnosis and treatment. Eur. Respir. Rev. 2016, 25, 472–486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walker, S.; Mercer, R.; Maskell, N.; Rahman, N.M. Malignant pleural effusion management: Keeping the flood gates shut. Lancet Respir. Med. 2019. [Google Scholar] [CrossRef]
- Davidson, B. Molecular testing on serous effusions. Diagn. Cytopathol. 2020. [Google Scholar] [CrossRef] [Green Version]
- Roy-Chowdhuri, S. Tumor-derived cell-free DNA in body cavity effusion supernatants: A promising alternative for genomic profiling. Cancer Cytopathol. 2020, 128, 14–16. [Google Scholar] [CrossRef] [Green Version]
- Pinto, D.; Schmitt, F. Current applications of molecular testing on body cavity fluids. Diagn. Cytopathol. 2020. [Google Scholar] [CrossRef]
- Huang, M.; Wei, S. Overview of Molecular Testing of Cytology Specimens. Acta Cytol. 2020, 64, 136–146. [Google Scholar] [CrossRef] [PubMed]
- Kalemkerian, G.P.; Narula, N.; Kennedy, E.B.; Biermann, W.A.; Donington, J.; Leighl, N.B.; Lew, M.; Pantelas, J.; Ramalingam, S.S.; Reck, M.; et al. Molecular testing guideline for the selection of patients with lung cancer for treatment with targeted tyrosine kinase inhibitors: American society of clinical oncology endorsement of the college of American pathologists/international association for the study of lung cancer/association for molecular pathology clinical practice guideline update. J. Clin. Oncol. 2018, 36, 911–919. [Google Scholar] [CrossRef] [PubMed]
- Lindeman, N.I.; Cagle, P.T.; Aisner, D.L.; Arcila, M.E.; Beasley, M.B.; Bernicker, E.H.; Colasacco, C.; Dacic, S.; Hirsch, F.R.; Kerr, K.; et al. Updated Molecular Testing Guideline for the Selection of Lung Cancer Patients for Treatment With Targeted Tyrosine Kinase Inhibitors: Guideline From the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. J. Thorac. Oncol. 2018, 13, 323–358. [Google Scholar] [PubMed] [Green Version]
- Yu, G.H.; Glaser, L.J.; Gustafson, K.S. Role of Ancillary Techniques in Fluid Cytology. Acta Cytol. 2020, 64, 52–62. [Google Scholar] [CrossRef] [PubMed]
- Roy-Chowdhuri, S.; Mehrotra, M.; Bolivar, A.M.; Kanagal-Shamanna, R.; Barkoh, B.A.; Hannigan, B.; Zalles, S.; Ye, W.; Duose, D.; Broaddus, R.; et al. Salvaging the supernatant: Next generation cytopathology for solid tumor mutation profiling. Mod. Pathol. 2018, 31, 1036–1045. [Google Scholar] [CrossRef] [Green Version]
- Malapelle, U.; Pisapia, P.; Rocco, D.; Smeraglio, R.; di Spirito, M.; Bellevicine, C.; Troncone, G. Next generation sequencing techniques in liquid biopsy: Focus on non-small cell lung cancer patients. Transl. Lung Cancer Res. 2016, 5, 505–510. [Google Scholar] [CrossRef] [Green Version]
- Aziz, N.; Zhao, Q.; Bry, L.; Driscoll, D.K.; Funke, B.; Gibson, J.S.; Grody, W.W.; Hegde, M.R.; Hoeltge, G.A.; Leonard, D.G.B.; et al. College of American Pathologists’ laboratory standards for next-generation sequencing clinical tests. Arch. Pathol. Lab. Med. 2015, 139, 481–493. [Google Scholar] [CrossRef] [Green Version]
- Luthra, R.; Chen, H.; Roy-Chowdhuri, S.; Singh, R.R. {Next-Generation} Sequencing in Clinical Molecular Diagnostics of Cancer: Advantages and Challenges. Cancers 2015, 7, 2023–2036. [Google Scholar] [CrossRef]
- Roy-Chowdhuri, S.; Pisapia, P.; Salto-Tellez, M.; Savic, S.; Nacchio, M.; de Biase, D.; Tallini, G.; Troncone, G.; Schmitt, F. Invited review-next-generation sequencing: A modern tool in cytopathology. Virchows Arch. 2019, 475, 3–11. [Google Scholar] [CrossRef]
- Jain, D.; Roy-Chowdhuri, S. Molecular Pathology of Lung Cancer Cytology Specimens: A Concise Review. Arch. Pathol. Lab. Med. 2018, 142, 1127–1133. [Google Scholar] [CrossRef] [Green Version]
- Hagemann, I.S.; Devarakonda, S.; Lockwood, C.M.; Spencer, D.H.; Guebert, K.; Bredemeyer, A.J.; Al-Kateb, H.; Nguyen, T.T.; Duncavage, E.J.; Cottrell, C.E.; et al. Clinical next-generation sequencing in patients with non–small cell lung cancer. Cancer 2015, 121, 631–639. [Google Scholar] [CrossRef] [PubMed]
- Vanderlaan, P.A.; Yamaguchi, N.; Folch, E.; Boucher, D.H.; Kent, M.S.; Gangadharan, S.P.; Majid, A.; Goldstein, M.A.; Huberman, M.S.; Kocher, O.N.; et al. Success and failure rates of tumor genotyping techniques in routine pathological samples with non-small-cell lung cancer. Lung Cancer 2014, 84, 39–44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mayo-de-las-Casas, C.; Garzón Ibáñez, M.; Jordana-Ariza, N.; García-Peláez, B.; Balada-Bel, A.; Villatoro, S.; Malapelle, U.; Karachaliou, N.; Troncone, G.; Rosell, R.; et al. An update on liquid biopsy analysis for diagnostic and monitoring applications in non-small cell lung cancer. Expert Rev. Mol. Diagn. 2018, 18, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Villatoro, S.; Mayo-de-las-Casas, C.; Jordana-Ariza, N.; Viteri-Ramírez, S.; Garzón-Ibañez, M.; Moya-Horno, I.; García-Peláez, B.; González-Cao, M.; Malapelle, U.; Balada-Bel, A.; et al. Prospective detection of mutations in cerebrospinal fluid, pleural effusion, and ascites of advanced cancer patients to guide treatment decisions. Mol. Oncol. 2019, 13, 2633–2645. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.; Luthra, R.; Goswami, R.S.; Singh, R.R.; Roy-Chowdhuri, S. Analysis of pre-analytic factors affecting the success of clinical next-generation sequencing of solid organ malignancies. Cancers 2015, 7, 1699–1715. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, J.; Hua, P.; Liu, N.; Li, Q.; Zhu, X.; Jiang, L.; Zheng, K.; Su, X. Targeted next-generation sequencing in cytology specimens for molecular profiling of lung adenocarcinoma. Int. J. Clin. Exp. Pathol. 2018, 11, 3647–3655. [Google Scholar]
- Yamamoto, G.; Kikuchi, M.; Kobayashi, S.; Arai, Y.; Fujiyoshi, K.; Wakatsuki, T.; Kakuta, M.; Yamane, Y.; Iijima, Y.; Mizutani, H.; et al. Routine genetic testing of lung cancer specimens derived from surgery, bronchoscopy and fluid aspiration by next generation sequencing. Int. J. Oncol. 2017, 50, 1579–1589. [Google Scholar] [CrossRef]
- Xiang, C.; Huo, M.; Ma, S.; Guo, L.; Zhao, R.; Teng, H.; Zhang, J.; Han, Y. Molecular Profiling for Supernatants and Matched Cell Pellets of Pleural Effusions in Non–Small-Cell Lung Cancer. J. Mol. Diagnostics 2020, 22, 513–522. [Google Scholar] [CrossRef]
- Liu, L.; Shao, D.; Deng, Q.; Tang, H.; Wang, J.; Liu, J.; Guo, F.; Lin, Y.; Peng, Z.; Mao, M.; et al. Next generation sequencing-based molecular profiling of lung adenocarcinoma using pleural effusion specimens. J. Thorac. Dis. 2018, 10, 2631–2637. [Google Scholar] [CrossRef]
- Zhang, P.; Wu, X.; Tang, M.; Nie, X.; Li, L. Detection of EGFR gene mutation status from pleural effusions and other body fluid specimens in patients with lung adenocarcinoma. Thorac. Cancer 2019, 10, 2218–2224. [Google Scholar] [CrossRef] [Green Version]
- Song, Z.; Cheng, G.; Zhang, Y. PD-L1 expression in malignant pleural effusion samples and its correlation with oncogene mutations in non-small cell lung cancer. J. Thorac. Dis. 2020, 12, 1385–1392. [Google Scholar] [CrossRef] [PubMed]
- Reck, M.; Rodríguez-Abreu, D.; Robinson, A.G.; Hui, R.; Csőszi, T.; Fülöp, A.; Gottfried, M.; Peled, N.; Tafreshi, A.; Cuffe, S.; et al. Pembrolizumab versus Chemotherapy for PD-L1–Positive Non–Small-Cell Lung Cancer. N. Engl. J. Med. 2016, 375, 1823–1833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.G.; Zeng, D.X.; Huang, J.A.; Jiang, J.H. Effective assessment of low times MET amplification in pleural effusion after epidermal growth factor receptor-tyrosine kinase inhibitors (EGFR-TKIs) acquired resistance. Medicine 2018, 97. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Ma, Z.; Zhang, Y.; Li, D.; Lv, D.; Chen, Z.; Li, P.; AI-Dherasi, A.; Zheng, F.; Tian, J.; et al. Targeted deep sequencing from multiple sources demonstrates increased NOTCH1 alterations in lung cancer patient plasma. Cancer Med. 2019, 8, 5673–5686. [Google Scholar] [CrossRef] [Green Version]
- Esagian, S.M.; Grigoriadou, G.; Nikas, I.P.; Boikou, V.; Sadow, P.M.; Won, J.K.; Economopoulos, K.P. Comparison of liquid-based to tissue-based biopsy analysis by targeted next generation sequencing in advanced non-small cell lung cancer: A comprehensive systematic review. J. Cancer Res. Clin. Oncol. 2020, 146, 2051–2066. [Google Scholar] [CrossRef]
- Cantey, E.P.; Walter, J.M.; Corbridge, T.; Barsuk, J.H. Complications of thoracentesis. Curr. Opin. Pulm. Med. 2016, 22, 378–385. [Google Scholar] [CrossRef]
- Tong, L.; Ding, N.; Tong, X.; Li, J.; Zhang, Y.; Wang, X.; Xu, X.; Ye, M.; Li, C.; Wu, X.; et al. Tumor-derived {DNA} from pleural effusion supernatant as a promising alternative to tumor tissue in genomic profiling of advanced lung cancer. Theranostics 2019, 9, 5532–5541. [Google Scholar] [CrossRef]
- Leichsenring, J.; Volckmar, A.L.; Kirchner, M.; Kazdal, D.; Kriegsmann, M.; Stögbauer, F.; Bockmayr, T.; Klauschen, F.; Herth, F.J.F.; Penzel, R.; et al. Targeted deep sequencing of effusion cytology samples is feasible, informs spatiotemporal tumor evolution, and has clinical and diagnostic utility. Genes Chromosom. Cancer 2018, 57, 70–79. [Google Scholar] [CrossRef]
- Yang, S.-R.R.; Mooney, K.L.; Libiran, P.; Jones, C.D.; Joshi, R.; Lau, H.D.; Stehr, H.; Berry, G.J.; Zehnder, J.L.; Long, S.R.; et al. Targeted deep sequencing of cell-free DNA in serous body cavity fluids with malignant, suspicious, and benign cytology. Cancer Cytopathol. 2020, 128, 43–56. [Google Scholar] [CrossRef]
- Yang, S.R.; Lin, C.Y.; Stehr, H.; Long, S.R.; Kong, C.S.; Berry, G.J.; Zehnder, J.L.; Kunder, C.A. Comprehensive Genomic Profiling of Malignant Effusions in Patients with Metastatic Lung Adenocarcinoma. J. Mol. Diagn. 2018, 20, 184–194. [Google Scholar] [CrossRef] [Green Version]
- Carter, J.; Miller, J.A.; Feller-Kopman, D.; Ettinger, D.; Sidransky, D.; Maleki, Z. Molecular profiling of malignant pleural effusion in metastatic non-small-cell lung carcinoma the effect of preanalytical factors. Ann. Am. Thorac. Soc. 2017, 14, 1169–1176. [Google Scholar] [CrossRef]
- Buttitta, F.; Felicioni, L.; Del Grammastro, M.; Filice, G.; Di Lorito, A.; Malatesta, S.; Viola, P.; Centi, I.; D’Antuono, T.; Zappacosta, R.; et al. Effective assessment of egfr mutation status in bronchoalveolar lavage and pleural fluids by next-generation sequencing. Clin. Cancer Res. 2013, 19, 691–698. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Li, X.; Wang, W.; Shao, Y.; Zhang, Y.; Song, Z. Gene Alterations in Paired Supernatants and Precipitates from Malignant Pleural Effusions of Non-Squamous Non-Small Cell Lung Cancer. Transl. Oncol. 2020, 13, 100784. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.; Lieberman, D.; Morrissette, J.J.D.; Baloch, Z.W.; Roth, D.B.; McGrath, C. Using “residual” FNA rinse and body fluid specimens for next-generation sequencing: An institutional experience. Cancer Cytopathol. 2016, 124, 324–329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, Z.; Cai, Z.; Yan, J.; Shao, Y.W.; Zhang, Y. Liquid biopsies using pleural effusion-derived exosomal DNA in advanced lung adenocarcinoma. Transl. Lung Cancer Res. 2019, 8, 392–400. [Google Scholar] [CrossRef] [PubMed]
- DiBardino, D.M.; Saqi, A.; Elvin, J.A.; Greenbowe, J.; Suh, J.H.; Miller, V.A.; Ali, S.M.; Stoopler, M.; Bulman, W.A. Yield and Clinical Utility of Next-Generation Sequencing in Selected Patients With Lung Adenocarcinoma. Clin. Lung Cancer 2016, 17, 517–522.e3. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, M.E.; Montesion, M.; Young, L.; Suh, J.; Greenbowe, J.; Kennedy, M.; Giaccone, G.; Akerley, W.L.; Dowlati, A.; Creelan, B.C.; et al. Multiple configurations of EGFR exon 20 resistance mutations after first- and third-generation EGFR TKI treatment affect treatment options in NSCLC. PLoS ONE 2018, 13, e0208097. [Google Scholar] [CrossRef]
- Li, W.; Zhang, Z.; Guo, L.; Qiu, T.; Ling, Y.; Cao, J.; Guo, H.; Zhao, H.; Li, L.; Ying, J. Assessment of cytology based molecular analysis to guide targeted therapy in advanced non-small-cell lung cancer. Oncotarget 2016, 7, 8332–8340. [Google Scholar] [CrossRef]
- Li, H.; Liu, Z.Y.; Wu, N.; Chen, Y.C.; Cheng, Q.; Wang, J. PARP inhibitor resistance: The underlying mechanisms and clinical implications. Mol. Cancer 2020, 19, 107. [Google Scholar] [CrossRef]
- Patterson, M.J.; Sutton, R.E.; Forrest, I.; Sharrock, R.; Lane, M.; Kaufmann, A.; O’Donnell, R.; Edmondson, R.J.; Wilson, B.T.; Curtin, N.J. Assessing the function of homologous recombination DNA repair in malignant pleural effusion (MPE) samples. Br. J. Cancer 2014, 111, 94–100. [Google Scholar] [CrossRef]
- Roscilli, G.; De Vitis, C.; Ferrara, F.F.; Noto, A.; Cherubini, E.; Ricci, A.; Mariotta, S.; Giarnieri, E.; Giovagnoli, M.R.; Torrisi, M.R.; et al. Human lung adenocarcinoma cell cultures derived from malignant pleural effusions as model system to predict patients chemosensitivity. J. Transl. Med. 2016, 14, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Xu, Y.M.; Zou, Y.Q.; Lin, J.; Huang, B.; Liu, J.; Li, J.; Zhang, J.; Yang, W.M.; Min, Q.H.; et al. Identification of differential expressed PE exosomal miRNA in lung adenocarcinoma, tuberculosis, and other benign lesions. Medicine 2017, 96. [Google Scholar] [CrossRef] [PubMed]
- Shah, R.H.; Scott, S.N.; Brannon, A.R.; Levine, D.A.; Lin, O.; Berger, M.F. Comprehensive mutation profiling by next-generation sequencing of effusion fluids from patients with high-grade serous ovarian carcinoma. Cancer Cytopathol. 2015, 123, 289–297. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.; Margol, A.S.; Shukla, A.; Ren, X.; Finlay, J.L.; Krieger, M.D.; Gilles, F.H.; Couch, F.J.; Aziz, M.; Fung, E.T.; et al. Disseminated medulloblastoma in a child with germline BRCA2 6174delT mutation and without Fanconi anemia. Front. Oncol. 2015, 5. [Google Scholar] [CrossRef] [Green Version]
- Zhou, S.; Xu, B.; Qi, L.; Zhu, D.; Liu, B.; Wei, J. Next-generation sequencing reveals mutational accordance between cell-free {DNA} from plasma, malignant pleural effusion and ascites and directs targeted therapy in a gastric cancer patient. Cancer Biol. Ther. 2019, 20, 15–20. [Google Scholar] [CrossRef]
- da Cunha Santos, G. Standardizing preanalytical variables for molecular cytopathology. Cancer Cytopathol. 2013, 121, 341–343. [Google Scholar] [CrossRef] [PubMed]
- Husain, H.; Nykin, D.; Bui, N.; Quan, D.; Gomez, G.; Woodward, B.; Venkatapathy, S.; Duttagupta, R.; Fung, E.; Lippman, S.M.; et al. {Cell-Free} {DNA} from Ascites and Pleural Effusions: Molecular Insights into Genomic Aberrations and Disease Biology. Mol. Cancer Ther. 2017, 16, 948–955. [Google Scholar] [CrossRef] [Green Version]
- Ye, W.; Hannigan, B.; Zalles, S.; Mehrotra, M.; Barkoh, B.A.; Williams, M.D.; Cabanillas, M.E.; Edeiken-Monroe, B.; Hu, P.; Duose, D.; et al. Centrifuged supernatants from {FNA} provide a liquid biopsy option for clinical next-generation sequencing of thyroid nodules. Cancer Cytopathol. 2019, 127, 146–160. [Google Scholar] [CrossRef] [Green Version]
- Hannigan, B.; Ye, W.; Mehrotra, M.; Lam, V.; Bolivar, A.; Zalles, S.; Barkoh, B.A.; Duose, D.; Hu, P.C.; Broaddus, R.; et al. Liquid biopsy assay for lung carcinoma using centrifuged supernatants from fine-needle aspiration specimens. Ann. Oncol. 2019, 30, 963–969. [Google Scholar] [CrossRef]
- Fernandes Marques, J.; Pereira Reis, J.; Fernandes, G.; Hespanhol, V.; Machado, J.C.; Costa, J.L. Circulating Tumor {DNA}: A Step into the Future of Cancer Management. Acta Cytol. 2019, 63, 456–465. [Google Scholar] [CrossRef]
- Martinez-Martin, N.; Magnus, D. Privacy and ethical challenges in next-generation sequencing. Expert Rev. Precis. Med. Drug Dev. 2019, 4, 95–104. [Google Scholar] [CrossRef] [PubMed]

| First Author/Reference | Pleural Fluid Material | Summary of Findings |
|---|---|---|
| Zhang et al. [26] | Cell blocks |
|
| ||
| ||
| Yamamoto et al. [27] | N/A |
|
| Xiang et al. [28] | Supernatants, Cell blocks |
|
| Liu et al. [29] | Cell blocks |
|
| Zhang et al. [30] | Supernatants, sDNA |
|
| ||
| Tong et al. [37] | Supernatants, sDNA |
|
| ||
| Villatoro et al. [24] | Supernatants |
|
| Liao et al. [34] | Supernatants, Cell pellets |
|
| ||
| ||
| Leichsenring et al. [38] | Cell blocks |
|
| Wang et al. [33] | N/A |
|
| ||
| Yang et al. [39] | Supernatants |
|
| First Author/Reference | Pleural Fluid Material | Summary of Findings |
|---|---|---|
| Yang et al. [39] | Supernatants |
|
| ||
| Yang et al. [40] | Cell blocks |
|
| Leichsenring et al. [38] | Cell blocks |
|
| Buttitta et al. [42] | Cell blocks |
|
| ||
| Liu et al. [29] | Cell blocks |
|
| Carter et al. [41] | Cell blocks |
|
| First Author/Reference | Pleural Fluid Material | Summary of Findings |
|---|---|---|
| Xiang et al. [28] | Supernatants, Cell blocks |
|
| ||
| ||
| Yang et al. [39] | Supernatants, Cell blocks |
|
| Li et al. [43] | Supernatants, Cell blocks |
|
| Wei et al. [44] | Aspirate, Cell blocks |
|
| Zhang et al. [30] | Supernatants, sDNA |
|
| ||
| Tong et al. [37] | Supernatants, sDNA |
|
| ||
| ||
| ||
| ||
| ||
| ||
| ||
| ||
| Song et al. [45] | Supernatants, Tumor exosomes |
|
| ||
| ||
| Liao et al. [34] | Supernatants, Cell pellets |
|
| First Author/Reference | Pleural Fluid Material | Summary of Findings |
|---|---|---|
| Leichserning et al. [38] | Cell blocks |
|
| DiBardino et al. [46] | Cell blocks, slides |
|
| Yang et al. [39] | Supernatants |
|
| Villatoro et al. [24] | Supernatants |
|
| Zhang et al. [26] | Cell blocks |
|
| Goldberg et al. [47] | N/A |
|
| Li et al. [48] | N/A |
|
| Wang et al. [52] | N/A |
|
| Tong et al. [37] | Supernatants, sDNA |
|
| Patterson et al. [50] | Cell cultures |
|
| Roscilli et al. [51] | Cell cultures from cell pellets |
|
| ||
| ||
| ||
| Song et al. [31] | Cell blocks |
|
| First Author/Reference | Cancer | Pleural Fluid Material | Summary of Findings |
|---|---|---|---|
| Yang et al. [39] | Lung, Breast, GI/Pancreas, Primary peritoneal | Supernatants |
|
| |||
| Shah et al. [53] | Ovarian | Cytospins |
|
| |||
| Xu et al. [54] | Medulloblastoma | N/A |
|
| Zhou et al. [55] | Gastric | Supernatants |
|
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grigoriadou, G.Ι.; Esagian, S.M.; Ryu, H.S.; Nikas, I.P. Molecular Profiling of Malignant Pleural Effusions with Next Generation Sequencing (NGS): Evidence that Supports Its Role in Cancer Management. J. Pers. Med. 2020, 10, 206. https://0-doi-org.brum.beds.ac.uk/10.3390/jpm10040206
Grigoriadou GΙ, Esagian SM, Ryu HS, Nikas IP. Molecular Profiling of Malignant Pleural Effusions with Next Generation Sequencing (NGS): Evidence that Supports Its Role in Cancer Management. Journal of Personalized Medicine. 2020; 10(4):206. https://0-doi-org.brum.beds.ac.uk/10.3390/jpm10040206
Chicago/Turabian StyleGrigoriadou, Georgia Ι., Stepan M. Esagian, Han Suk Ryu, and Ilias P. Nikas. 2020. "Molecular Profiling of Malignant Pleural Effusions with Next Generation Sequencing (NGS): Evidence that Supports Its Role in Cancer Management" Journal of Personalized Medicine 10, no. 4: 206. https://0-doi-org.brum.beds.ac.uk/10.3390/jpm10040206