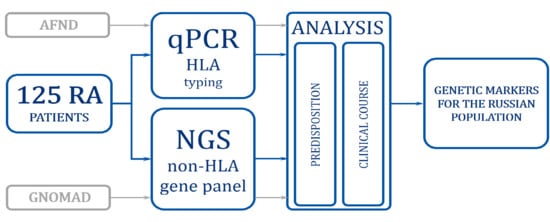
Genetic Factors of Predisposition and Clinical Characteristics of Rheumatoid Arthritis in Russian Patients
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethical Approval and Patients
2.2. DNA Isolation and Genotyping HLA and Non-HLA Genes
2.3. Statistical Analysis
3. Results
3.1. Identification of Genetic Factors of Predisposition to RA
3.2. Genetic Factors Associated with the Clinical Course of the Disease
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO. Available online: http://www.who.int/chp/topics/rheumatic/en/ (accessed on 4 December 2012).
- van Vollenhoven, R.F. Sex Differences in Rheumatoid Arthritis: More than Meets the Eye. BMC Med. 2009, 7, 12. [Google Scholar] [CrossRef] [Green Version]
- Deane, K.D.; Demoruelle, M.K.; Kelmenson, L.B.; Kuhn, K.A.; Norris, J.M.; Holers, V.M. Genetic and Environmental Risk Factors for Rheumatoid Arthritis. Best Pract. Res. Clin. Rheumatol. 2017, 3–18. [Google Scholar] [CrossRef]
- Karami, J.; Aslani, S.; Jamshidi, A.; Garshasbi, M.; Mahmoudi, M. Genetic Implications in the Pathogenesis of Rheumatoid Arthritis: An Updated Review. Gene 2019, 8–16. [Google Scholar] [CrossRef]
- Suzuki, A.; Terao, C.; Yamamoto, K. Linking of Genetic Risk Variants to Disease-Specific Gene Expression via Multi-Omics Studies in Rheumatoid Arthritis. Semin. Arthritis Rheum. 2019, 49, S49–S53. [Google Scholar] [CrossRef]
- Mikhaylenko, D.S.; Nemtsova, M.V.; Bure, I.V.; Kuznetsova, E.B.; Alekseeva, E.A.; Tarasov, V.V.; Lukashev, A.N.; Beloukhova, M.I.; Deviatkin, A.A.; Zamyatnin, A.A. Genetic Polymorphisms Associated with Rheumatoid Arthritis Development and Antirheumatic Therapy Response. Int. J. Mol. Sci. 2020, 21, 4911. [Google Scholar] [CrossRef]
- Yamamoto, K.; Okada, Y.; Suzuki, A.; Kochi, Y. Genetics of Rheumatoid Arthritis in Asia—Present and Future. Nature Reviews Rheumatology; Nature Publishing Group: Berlin, Germany, 2015; pp. 375–379. [Google Scholar] [CrossRef]
- Kuzhir, T. Polygenic basis of the rheumatoid arthritis. Ecol. Genet. 2019, 17, 77–90. [Google Scholar] [CrossRef] [Green Version]
- Yatskiu, H.A.; Zlotnikova, M.V.; Sukalo, A.V.; Goncharova, R.I. Characteristic Spectra of Class I and II HLA-Alleles in Patients with Different Clinical Forms of Juvenile Idiopathic Arthritis in the Republic of Belarus. Dokl. Natl. Acad. Sci. Belarus 2020, 64, 209–216. [Google Scholar] [CrossRef]
- Bogunia-Kubik, K.; Świerkot, J.; Malak, A.; Wysoczańska, B.; Nowak, B.; Białowąs, K.; Gębura, K.; Korman, L.; Wiland, P. IL-17A, IL-17F and IL-23R Gene Polymorphisms in Polish Patients with Rheumatoid Arthritis. Arch. Immunol. Ther. Exp. (Warsz.) 2015, 63, 215–221. [Google Scholar] [CrossRef] [Green Version]
- Mikhaylenko, D.S.; Nemtsova, M.V.; Bure, I.V.; Kuznetsova, E.B.; Alekseeva, E.A.; Deviatkin, A.A.; Zamyatnin, A.A. IAD177464_185_coverage_summary. Csv. Figshare. Dataset. 2021. [Google Scholar] [CrossRef]
- AFND. Available online: http://www.allelefrequencies.net/population.asp?pop_id=3322 (accessed on 16 June 2015).
- gnomAD. Available online: https://gnomad.broadinstitute.org/ (accessed on 17 October 2018).
- Machaj, F.; Rosik, J.; Szostak, B.; Pawlik, A. The Evolution in Our Understanding of the Genetics of Rheumatoid Arthritis and the Impact on Novel Drug Discovery. Expert Opin. Drug Discov. 2020, 15, 85–99. [Google Scholar] [CrossRef]
- Guseva, I.A.; Demidova, N.Y.; Soroka, N.E.; Novikov, A.A.; Lutchihina, E.L.; Alexandrova, E.N.; Lukina, G.V.; Fedorenko, E.V.; Aronova, E.S.; Samarkina, E.Y.; et al. Inununogenetic Aspects of Early Rheumatoid Arthritis. Vestn. Ross. Akad. Meditsinskikh Nauk 2013, 68, 36–43. [Google Scholar] [CrossRef] [Green Version]
- Kuranov, A.B.; Boldyreva, M.N.; Momynaliev, K.T. A Meta-Analysis of Associations of HLA-DRB1 with Rheumatoid Arthritis. Immunologiya 2017, 38, 140–143. [Google Scholar] [CrossRef]
- Okada, Y.; Kim, K.; Han, B.; Pillai, N.E.; Ong, R.T.H.; Saw, W.Y.; Luo, M.; Jiang, L.; Yin, J.; Bang, S.Y.; et al. Risk for ACPA-Positive Rheumatoid Arthritis is Driven by Shared HLA Amino Acid Polymorphisms in Asian and European Populations. Hum. Mol. Genet. 2014, 23, 6916–6926. [Google Scholar] [CrossRef] [Green Version]
- Bettencourt, A.; Carvalho, C.; Leal, B.; Brás, S.; Lopes, D.; Martins Da Silva, A.; Santos, E.; Torres, T.; Almeida, I.; Farinha, F.; et al. The Protective Role of HLA-DRB1*13 in Autoimmune Diseases. J. Immunol. Res. 2015, 2015. [Google Scholar] [CrossRef]
- Van Der Woude, D.; Lie, B.A.; Lundström, E.; Balsa, A.; Feitsma, A.L.; Houwing-Duistermaat, J.J.; Verduijn, W.; Nordang, G.B.N.; Alfredsson, L.; Klareskog, L.; et al. Protection against Anti-Citrullinated Protein Antibody-Positive Rheumatoid Arthritis Is Predominantly Associated with HLA-DRB1*1301: A Meta-Analysis of HLA-DRB1 Associations with Anti-Citrullinated Protein Antibody-Positive and Anti-Citrullinated Protein. Arthritis Rheum. 2010, 62, 1236–1245. [Google Scholar] [CrossRef]
- Fiorillo, E.; Orrú, V.; Stanford, S.M.; Liu, Y.; Salek, M.; Rapini, N.; Schenone, A.D.; Saccucci, P.; Delogu, L.G.; Angelini, F.; et al. Autoimmune-Associated PTPN22 R620W Variation Reduces Phosphorylation of Lymphoid Phosphatase on an Inhibitory Tyrosine Residue. J. Biol. Chem. 2010, 285, 26506–26518. [Google Scholar] [CrossRef] [Green Version]
- Abbasifard, M.; Imani, D.; Bagheri-Hosseinabadi, Z. PTPN22 Gene Polymorphism and Susceptibility to Rheumatoid Arthritis (RA): Updated Systematic Review and Meta-Analysis. J. Gene Med. 2020. [Google Scholar] [CrossRef]
- Gregersen, P.K.; Lee, H.S.; Batliwalla, F.; Begovich, A.B. PTPN22: Setting Thresholds for Autoimmunity. Seminars in Immunology. Semin. Immunol. 2006, 214–223. [Google Scholar] [CrossRef]
- Mihailova, A.; Mikazane, H.; Klovins, J.; Nikitina-Zake, L. Association of Protein Tyrosine Phosphatase Non-Receptor 22 (PTPN22) Rs2476601 and Kruppel-like Factor 12 (KLF12) Rs1324913 Single Nucleotide Polymorphisms with Rheumatoid Arthritis in a Latvian Population. Scand. J. Rheumatol. 2011, 491–492. [Google Scholar] [CrossRef]
- Song, G.G.; Bae, S.-C.; Kim, J.-H.; Lee, Y.H. The PTPN22 C1858T Polymorphism and Rheumatoid Arthritis: A Meta-Analysis. Rheumatol. Int. 2013, 33, 1991–1999. [Google Scholar] [CrossRef]
- Lamana, A.; López-Santalla, M.; Castillo-González, R.; Ortiz, A.M.; Martín, J.; García-Vicuña, R.; González-Álvaro, I. The Minor Allele of Rs7574865 in the STAT4 Gene Is Associated with Increased MRNA and Protein Expression. PLoS ONE 2015, 10, e0142683. [Google Scholar] [CrossRef]
- Kariuki, S.N.; Kirou, K.A.; MacDermott, E.J.; Barillas-Arias, L.; Crow, M.K.; Niewold, T.B. Cutting Edge: Autoimmune Disease Risk Variant of STAT4 Confers Increased Sensitivity to IFN-α in Lupus Patients In Vivo. J. Immunol. 2009, 182, 34–38. [Google Scholar] [CrossRef] [Green Version]
- Stark, K.; Rovenský, J.; Blažičková, S.; Grosse-Wilde, H.; Ferencik, S.; Hengstenberg, C.; Straub, R.H. Association of Common Polymorphisms in Known Susceptibility Genes with Rheumatoid Arthritis in a Slovak Population Using Osteoarthritis Patients as Controls. Arthritis Res. Ther. 2009, 11. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Y.; Zhang, R.; Zheng, J.; Liu, P.; Tang, G.; Lv, H.; Zhang, L.; Shang, Z.; Zhan, Y.; Lv, W.; et al. Meta-Analysis of 125 Rheumatoid Arthritis-Related Single Nucleotide Polymorphisms Studied in the Past Two Decades. PLoS ONE 2012, 7, e51571. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.H.; Bae, S.C. Associations between FCGR2A Rs1801274, FCGR3A Rs396991, FCGR3B NA1/NA2 Polymorphisms and Periodontitis: A Meta-Analysis. Mol. Biol. Rep. 2013, 40, 4985–4993. [Google Scholar] [CrossRef]
- Kyogoku, C.; Tsuchiya, N.; Matsuta, K.; Tokunaga, K. Studies on the Association of Fcγ Receptor IIA, IIB, IIIA and IIIB Polymorphisms with Rheumatoid Arthritis in the Japanese: Evidence for a Genetic Interaction between HLA-DRB1 and FCGR3A. Genes Immun. 2002, 3, 488–493. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.-K.; Liu, J.; Liu, J.; Liang, Y.; Xu, W.-D.; Leng, R.-X.; Pan, H.-F.; Ye, D.-Q. Associations between PADI4 Gene Polymorphisms and Rheumatoid Arthritis: An Updated Meta-Analysis. Arch. Med. Res. 2015, 46, 317–325. [Google Scholar] [CrossRef]
- Rashid, T.; Ebringer, A. Autoimmunity in Rheumatic Diseases Is Induced by Microbial Infections via Crossreactivity or Molecular Mimicry. Autoimmune Dis. 2012. [Google Scholar] [CrossRef] [Green Version]
- Pawlik, A.; Wrzesniewska, J.; Florczak, M.; Gawronska-Szklarz, B.; Herczynska, M. The −590 IL-4 Promoter Polymorphism in Patients with Rheumatoid Arthritis. Rheumatol. Int. 2005, 26, 48–51. [Google Scholar] [CrossRef]
- Mohamed, F.Z.; Mussien, Y.M.; Basiony, E.F. Interleukin-4 (IL-4) and Interleukin-4 Receptor Alpha Chain (IL-4Rα) Gene Polymorphisms in Egyptian Rheumatic Arthritis Patients. Biochem. Lett. 2010, 6, 1–21. [Google Scholar] [CrossRef]
- Hussein, Y.; El-Tarhouny, S.; Mohamed, R.; Pasha, H.; Abul-Saoud, A. Association of Interleukin-4 Receptor Gene Polymorphisms with Rheumatoid Arthritis in Egyptian Female Patients. Jt. Bone Spine 2012, 79, 38–42. [Google Scholar] [CrossRef] [PubMed]
- Burgos, P.I.; Causey, Z.L.; Tamhane, A.; Kelley, J.M.; Brown, E.E.; Hughes, L.B.; Danila, M.I.; van Everdingen, A.; Conn, D.L.; Jonas, B.L.; et al. Association of IL4R Single-Nucleotide Polymorphisms with Rheumatoid Nodules in African Americans with Rheumatoid Arthritis. Arthritis Res. Ther. 2010, 12, R75. [Google Scholar] [CrossRef] [Green Version]
- Marinou, I.; Till, S.H.; Moore, D.J.; Wilson, A.G. Lack of Association or Interactions between the IL-4, IL-4Rα and IL-13 Genes, and Rheumatoid Arthritis. Arthritis Res. Ther. 2008, 10, R80. [Google Scholar] [CrossRef] [Green Version]
- Nell, V.P.K.; Machold, K.P.; Stamm, T.A.; Eberl, G.; Heinzl, H.; Uffmann, M.; Smolen, J.S.; Steiner, G. Autoantibody Profiling as Early Diagnostic and Prognostic Tool for Rheumatoid Arthritis. Ann. Rheum. Dis. 2005, 64, 1731–1736. [Google Scholar] [CrossRef]
- Shmerling, R.H.; Delbanco, T.L. The Rheumatoid Factor: An Analysis of Clinical Utility. Am. J. Med. 1991, 91, 528–534. [Google Scholar] [CrossRef]
- Terao, C.; Ohmura, K.; Kochi, Y.; Ikari, K.; Okada, Y.; Shimizu, M.; Nishina, N.; Suzuki, A.; Myouzen, K.; Kawaguchi, T.; et al. Anti-Citrullinated Peptide/Protein Antibody (ACPA)-Negative RA Shares a Large Proportion of Susceptibility Loci with ACPA-Positive RA: A Meta-Analysis of Genome-Wide Association Study in a Japanese Population. Arthritis Res. Ther. 2015, 17. [Google Scholar] [CrossRef] [Green Version]
- Balandraud, N.; Picard, C.; Reviron, D.; Landais, C.; Toussirot, E.; Lambert, N.; Telle, E.; Charpin, C.; Wendling, D.; Pardoux, E.; et al. HLA-DRB1 Genotypes and the Risk of Developing Anti Citrullinated Protein Antibody (ACPA) Positive Rheumatoid Arthritis. PLoS ONE 2013, 8, e64108. [Google Scholar] [CrossRef] [Green Version]
- Donlin, L.T.; Park, S.H.; Giannopoulou, E.; Ivovic, A.; Park-Min, K.H.; Siegel, R.M.; Ivashkiv, L.B. Insights into Rheumatic Diseases from Next-Generation Sequencing. Nat. Rev. Rheumatol. 2019, 327–339. [Google Scholar] [CrossRef]

| Clinical Characteristic | RA (N = 125) |
|---|---|
| Age (years) | |
| mean ± SD | 50.4 ± 13.14 |
| [Min, max] | [22, 82] |
| Gender | |
| Female | 109 (87.2%) |
| Male | 16 (12.8%) |
| Weight (kg) | |
| mean ± SD | 73.63 ± 16.366 |
| [Min, max] | [41.0, 131.0] |
| Disease duration (years) | 5.91 |
| Min, max | [2.61, 11.16] |
| Disease severity | |
| Moderate (DAS28-CRP > 3.2 to ≤5.1) | 10 (8.0%) |
| High (DAS28-CRP > 5.1) | 113 (90.4%) |
| DAS28-CRP | |
| mean ± SD | 5.94 ± 0.643 |
| [Min, max] | [4.5, 8.1] |
| CDAI | |
| mean ± SD | 39.43 ± 8.701 |
| [Min, max] | [24.8, 69.3] |
| HAQ-DI | |
| mean ± SD | 1.6890 ± 0.4998 |
| [Min, max] | [0.125, 2.875] |
| CRP (mg/mL) | |
| mean ± SD | 21.0 ± 20.83 |
| [Min, max] | [1, 120] |
| Anti-CCP (IU/mL) | |
| mean ± SD | 664.35 ± 999.308 |
| [Min, max] | [0.4, 6044.8] |
| RF (IU/mL) | |
| mean ± SD | 192.2 ± 240.95 |
| [Min, max] | [7, 1540] |
| Anti-CCP (>10 IU/mL) | |
| Yes | 101 (80.8%) |
| No | 22 (17.6%) |
| RF (≥15 IU/mL) | |
| Yes | 105 (84.0%) |
| No | 20 (16.0%) |
| Basal Anti-CCP and RF | |
| Low | 25 (20.0%) |
| Medium | 81 (64.8%) |
| High | 17 (13.6%) |
| Non-HLA | Frequency of Alleles and Genotypes | p | OR [CI 95%] | |
|---|---|---|---|---|
| Alleles and Genotypes | (abs. Value/Frequency) | |||
| RA (n = 125) | Control (GnomAD) | |||
| PTPN22 (rs2476601) | ||||
| Allele C | 192/0.77 | 13661/0.89 | 0.0001 | 0.4 [0.3–0.57] |
| Allele T | 58/0.23 | 1753/0.11 | 2.3 [1.7–3.2] | |
| CC | 80/0.64 | 6058/0.79 | 0.001 | 0.5 [0.3–0.7] |
| CT | 32/0.26 | 1545/0.20 | 1.37 [0.9–2] | |
| TT | 13/0.1 | 104/0.013 | 8.5 [4.6–15.56] | |
| FCGR2A (rs1801274) | ||||
| Allele G | 105/0.42 | 7710/0.5 | 0.011 | 0.72 [0.56–0.93] |
| Allele A | 145/0.58 | 7668/0.5 | 1.4 [1.08–1.8] | |
| GG | 19/0.15 | 1959/0.255 | 0.025 | 0.52 [0.32–0.86] |
| GA | 67/0.54 | 3792/0.493 | 1.2 [0.83–1.7] | |
| AA | 39/0.31 | 1938/0.252 | 1.3 [0.92–1.97] | |
| STAT4 (rs7574865) | 0.025 | |||
| Allele G | 181/0.72 | 12071/0.78 | 0.72 [0.55–0.96] | |
| Allele T | 69/0.28 | 3331/0.22 | 1.4 [1.04–1.83] | |
| 0.068 | ||||
| GG | 64/0.52 | 4717/0.62 | 0.66 [0.5–0.94] | |
| GT | 53/0.42 | 2637/0.34 | 1.4 [0.99–2] | |
| TT | 8/0.06 | 347/0.04 | 1.45 [0.7–3] | |
| IL4 (rs2243250) | 0.0025 | |||
| Allele T | 63/0.25 | 2705/0.18 | 1.6 [1.2–2.1] | |
| Allele C | 187/0.75 | 12699/0.82 | 0.63 [0.47–0.84] | |
| 0.002 | ||||
| TT | 9/0.07 | 265/0.0344 | 2.2 [1.1–4.3] | |
| CT | 45/0.36 | 2175/0.2824 | 1.43 [0.99–2.07] | |
| CC | 71/0.57 | 5262/0.6832 | 0.61 [0.43–0.87] | |
| IL17A (rs2275913) | 0.034 | |||
| Allele G | 143/0.57 | 9822/0.64 | 0.76 [0.59–0.97] | |
| Allele A | 107/0.43 | 5574/0.36 | 1.32 [1.02–1.7] | |
| 0.015 | ||||
| GG | 45/0.36 | 3113/0.40 | 0.83 [0.57–1.2] | |
| GA | 53/0.42 | 3596/0.47 | 0.84 [0.59–1.2] | |
| AA | 27/0.22 | 989/0.13 | 1.87 [1.2–2.88] | |
| TPMT (rs2842934) | 0.002 | |||
| Allele A | 175/0.7 | 12074/0.78 | 0.64 [0.5–0.85] | |
| Allele G | 75/0.3 | 3330/0.22 | 1.55 [1.2–2] | |
| 0.0016 | ||||
| AA | 63/0.5 | 4714/0.61 | 0.64 [0.45–0.92] | |
| AG | 49/0.39 | 2646/0.34 | 1.23 [0.86–1.77] | |
| GG | 13/0.1 | 342/0.04 | 2.5 [1.4–4.5] | |
| TNF (rs1800629) | 0.0001 | |||
| Allele A | 21/0.08 | 2636/0.17 | 0.4 [0.3–0.7] | |
| Allele G | 229/0.92 | 12780/0.83 | 2.25 [1.4–3.5] | |
| N/A | ||||
| AA | 0/0 | 257/0.033 | 0 [0] | |
| GA | 21/0.17 | 2122/0.275 | 0.53 [0.33–0.85] | |
| GG | 104/0.83 | 5329/0.691 | 2.2 [1.4–3.54] | |
| BTNL2 (rs3817963) | 0.017 | |||
| Allele C | 82/0.33 | 4001/0.26 | 1.4 [1.06–1.8] | |
| Allele T | 168/0.67 | 11387/0.74 | 0.72 [0.55–0.94] | |
| 0.015 | ||||
| CC | 17/0.14 | 544/0.07 | 2 [1.23–3.5] | |
| CT | 48/0.38 | 2913/0.38 | 1.02 [0.7–1.5] | |
| TT | 60/0.48 | 4237/0.55 | 0.75 [0.5–1.07] | |
| HLA-alleles and genotypes | RA (n = 114) | Control (AFND) | p | OR [CI 95%] |
| HLA-DRB1 | ||||
| Allele *01 | 45/19.74 | 474/11.85 | 0.0004 | 4.7 [3.3–6.8] |
| Allele *04 | 62/27.19 | 427/10.675 | 0.00001 | 3.1 [2.3–4.26] |
| Allele *07 | 20/8.77 | 562/14.05 | 0.02 | 0.6 [0.4–0.9] |
| Allele *08 | 0/0 | 139/3.475 | N/A | N/A |
| Allele *13 | 18/7.89 | 548/13.7 | 0.01 | 0.5 [0.3–0.89] |
| *01:03 | 5/4.39 | 33/1.65 | 0.05 | 2.7 [1.05–7.1] |
| *01:04 | 7/6.14 | 49/2.45 | 0.017 | 2.6 [1.15–5.9] |
| *01:16 | 8/7.02 | 18/0.9 | 0.00001 | 8.3 [3.53–19.55] |
| *04:04 | 13/11.4 | 19/0.95 | 0.00001 | 13.4 [6.4–27.9] |
| *04:16 | 5/4.39 | 21/1.05 | 0.007 | 4.3 [1.6–11.7] |
| HLA-B27 | 0.001 | |||
| B27+ | 21/17.8 | 176/8.8 | 2.2 [1.37–3.7] | |
| B27− | 97/82.2 | 1824/91.2 | 0.45 [0.27–0.73] | |
| Alleles | p | Adj. p | Genotypes | p | Adj. p |
|---|---|---|---|---|---|
| HLA-DRB1*04 | 0.00001 | 0.0008 | HLA-DRB1*04:04 | 0.00001 | 0.001 |
| PTPN22(rs2476601) | 0.0001 | 0.004 | HLA-DRB1*01:16 | 0.00001 | 0.001 |
| TNF (rs1800629) | 0.0001 | 0.003 | PTPN22(rs2476601) | 0.001 | 0.038 |
| HLA-DRB1*01 | 0.0004 | 0.008 | TPMT (rs2842934) | 0.002 | 0.046 |
| HLA-B27 | 0.001 | 0.015 | B27 + DRB1*01:16 | 0.002 | 0.077 |
| TPMT (rs2842934) | 0.002 | 0.026 | B27 + DRB1*01:03 | 0.004 | 0.115 |
| IL4 (rs2243250) | 0.0025 | 0.028 | HLA-DRB1*04:16 | 0.007 | 0.16 |
| HLA-DRB1*13 | 0.01 | 0.098 | IL4 (rs2243250) | 0.007 | 0.13 |
| FCGR2A (rs1801274) | 0.011 | 0.095 | BTNL2 (rs3817963) | 0.015 | 0.24 |
| BTNL2 (rs3817963) | 0.017 | 0.13 | IL17A (rs2275913) | 0.015 | 0.22 |
| HLA-DRB1*07 | 0.02 | 0.14 | HLA-DRB1*01:04 | 0.017 | 0.22 |
| STAT4 (rs7574865) | 0.025 | 0.16 | FCGR2A (rs1801274) | 0.026 | 0.29 |
| IL17A (rs2275913) | 0.034 | 0.2 | B27 + DRB1*04:16 | 0.012 | 0.23 |
| STAT4 (rs7574865) | 0.068 | 0.78 |
| Gene | rs | p-Value | Associations with Baseline Value |
|---|---|---|---|
| DHODH | rs3213422 | 0.041 | CRP |
| MTHFR | rs180113 | 0.03 | |
| PADI4 | rs11203367 | 0.009 | CDAI |
| 0.012 | DAS28 | ||
| 0.007 | HAQ-DI | ||
| rs2240340 | |||
| rs11203366 | |||
| 0.012 | |||
| 0.02 | |||
| rs874881 | 0.014 | ||
| 0.017 | |||
| 0.016 | |||
| 0.003 | |||
| 0.043 | HAQ-DI | ||
| AMPD1 | rs17602729 | 0.037 | HAQ-DI |
| HLA-B27 | - | 0.008 | Anti-CCP |
| IL23R | rs7539625 | 0.014 | |
| rs10889671 | 0.028 | ||
| 0.023 | DAS28 | ||
| 0.005 | Anti-CCP | ||
| rs7530511 | 0.018 | HAQ-DI | |
| rs11465770 | |||
| IL2RB | rs3218253 | 0.022 | Anti-CCP |
| IL6R | rs2228144 | 0.043 | |
| STAT4 | rs7574865 | 0.037 | |
| TNFRSF1A | rs767455 | 0.022 | |
| rs1800693 | 0.01 | ||
| IL2RA | rs2104286 | 0.042 | RF |
| IRAK3 | rs11541076 | 0.011 | |
| IL4R | rs1801275 | 0.027 | |
| 0.002 | DAS28 | ||
| 0.041 | CDAI | ||
| IL4 | rs2243250 | 0.014 | CDAI |
| 0.015 | DAS28 | ||
| IL6 | rs2069849 | 0.032 | CDAI |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vetchinkina, E.A.; Mikhaylenko, D.S.; Kuznetsova, E.B.; Deryagina, T.A.; Alekseeva, E.A.; Bure, I.V.; Zamyatnin, A.A., Jr.; Nemtsova, M.V. Genetic Factors of Predisposition and Clinical Characteristics of Rheumatoid Arthritis in Russian Patients. J. Pers. Med. 2021, 11, 469. https://0-doi-org.brum.beds.ac.uk/10.3390/jpm11060469
Vetchinkina EA, Mikhaylenko DS, Kuznetsova EB, Deryagina TA, Alekseeva EA, Bure IV, Zamyatnin AA Jr., Nemtsova MV. Genetic Factors of Predisposition and Clinical Characteristics of Rheumatoid Arthritis in Russian Patients. Journal of Personalized Medicine. 2021; 11(6):469. https://0-doi-org.brum.beds.ac.uk/10.3390/jpm11060469
Chicago/Turabian StyleVetchinkina, Ekaterina A., Dmitry S. Mikhaylenko, Ekaterina B. Kuznetsova, Tatiana A. Deryagina, Ekaterina A. Alekseeva, Irina V. Bure, Andrey A. Zamyatnin, Jr., and Marina V. Nemtsova. 2021. "Genetic Factors of Predisposition and Clinical Characteristics of Rheumatoid Arthritis in Russian Patients" Journal of Personalized Medicine 11, no. 6: 469. https://0-doi-org.brum.beds.ac.uk/10.3390/jpm11060469