The Effects of Metformin on the Gut Microbiota of Patients with Type 2 Diabetes: A Two-Center, Quasi-Experimental Study
Abstract
:1. Introduction
2. Results
2.1. Differences between before and Four Weeks after Medication
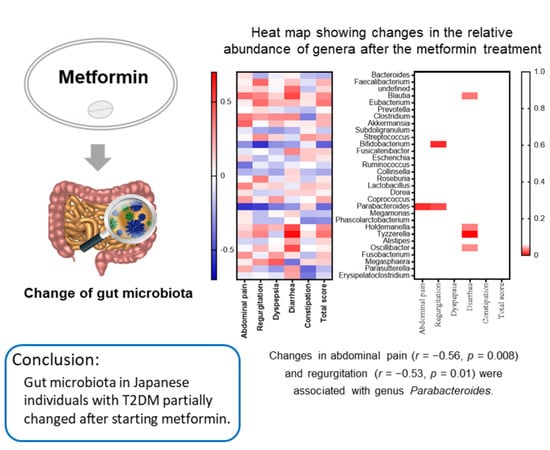
2.2. Subanalysis Excluding Medications that Affect Gut Microbiota
2.3. Relationships between the GSRS and Gut Microbiota in the Subanalysis
3. Discussion
4. Materials and Methods
4.1. Study Population
4.2. Ethical Considerations
4.3. Data Collection and Variables
4.4. Bacterial DNA Extraction From Feces and DNA Sequence Analysis
4.5. Microbiota Analysis
4.6. Statistical Analysis
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| T2DM | Type 2 diabetes mellitus |
| GSRS | Gastrointestinal Symptom Rating Scale |
| CVD | Cardiovascular disease |
| GLP-1 | Glucagon-like peptide 1 |
| GI | Gastrointestinal |
| QOL | Quality of life |
| BMI | Body mass index |
| HbA1c | Hemoglobin A1c |
| OTU | Operational taxonomic unit |
| ACE | Abundance-based coverage estimator |
| PCoA | Principal coordinates analysis |
| PERMANOVA | Permutational multivariate analysis of variance |
| WAD | Weighted average distance |
| LEfSe | Linear discriminant effect size |
| MAPK | Mitogen-activated protein kinase |
| SCFAs | Short-chain fatty acids |
| eGFR | Estimated glomerular filtration rate |
| PPI | Proton pump inhibitor |
| OD | Optical Density |
| QIIME | Quantitative Insights into Microbial Ecology |
| LDA | Linear discriminant analysis |
References
- Roglic, G.; World Health Organization. Global Report on Diabetes; World Health Organization: Geneva, Switzerland, 2016. [Google Scholar]
- Sato, J.; Kanazawa, A.; Ikeda, F.; Yoshihara, T.; Goto, H.; Abe, H.; Komiya, K.; Kawaguchi, M.; Shimizu, T.; Ogihara, T.; et al. Gut dysbiosis and detection of “Live gut bacteria” in blood of Japanese patients with type 2 diabetes. Diabetes Care 2014, 37, 2343–2350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kyriachenko, Y.; Falalyeyeva, T.; Korotkyi, O.; Molochek, N.; Kobyliak, N. Crosstalk between gut microbiota and antidiabetic drug action. World J. Diabetes 2019, 10, 154–168. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Wang, M.; Yang, J.; Xu, Q.; Liang, C.; Chen, B.; Zhang, J.; Yang, Y.; Wang, H.; Shang, Y.; et al. Response of gut microbiota in type 2 diabetes to hypoglycemic agents. Endocrine 2019, 66, 485–493. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, Y.; Hamaguchi, M.; Kaji, A.; Sakai, R.; Osaka, T.; Inoue, R.; Kashiwagi, S.; Mizushima, K.; Uchiyama, K.; Takagi, T.; et al. Intake of sucrose affects gut dysbiosis in patients with type 2 diabetes. J. Diabetes Investig. 2020. [Google Scholar] [CrossRef] [PubMed]
- Olivares, M.; Neyrinck, A.; Pötgens, S.; Beaumont, M.; Salazar, N.; Cani, P.; Bindels, L.; Delzenne, N. The DPP-4 inhibitor vildagliptin impacts the gut microbiota and prevents disruption of intestinal homeostasis induced by a Western diet in mice. Diabetologia 2018, 61, 1838–1848. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nathan, D.; Buse, J.; Davidson, M.; Ferrannini, E.; Holman, R.; Sherwin, R.; Zinman, B. Medical management of hyperglycemia in type 2 diabetes: A consensus algorithm for the initiation and adjustment of therapy. Diabetes Care 2009, 32, 193–203. [Google Scholar] [CrossRef] [Green Version]
- Pernicova, I.; Korbonits, M. Metformin-Mode of action and clinical implications for diabetes and cancer. Nat. Rev. Endocrinol. 2014, 10, 143–156. [Google Scholar] [CrossRef]
- Duca, F.A.; Côté, C.; Rasmussen, B.; Zadeh-Tahmasebi, M.; Rutter, G.; Filippi, B.; Lam, T. Metformin activates a duodenal Ampk-dependent pathway to lower hepatic glucose production in rats. Nat. Med. 2015, 21, 506–511. [Google Scholar] [CrossRef]
- McCreight, L.; Bailey, C.; Pearson, E. Metformin and the gastrointestinal tract. Diabetologia 2016, 59, 426–435. [Google Scholar] [CrossRef] [Green Version]
- Bryrup, T.; Thomsen, C.; Kern, T.; Allin, K.; Brandslund, I.; Jørgensen, N.; Vestergaard, H.; Hansen, T.; Hansen, T.H.; Pedersen, O.; et al. Metformin-induced changes of the gut microbiota in healthy young men: Results of a non-blinded, one-armed intervention study. Diabetologia 2019, 62, 1024–1035. [Google Scholar] [CrossRef] [Green Version]
- Wu, H.; Esteve, E.; Tremaroli, V.; Khan, M.; Caesar, R.; Mannerås-Holm, L.; Ståhlman, M.; Olsson, L.; Serino, M.; Planas-Fèlix, M.; et al. Metformin alters the gut microbiome of individuals with treatment-naive type 2 diabetes, contributing to the therapeutic effects of the drug. Nat. Med. 2017, 23, 850–858. [Google Scholar] [CrossRef] [PubMed]
- Cuesta-Zuluaga, J.; Mueller, N.; Corrales-Agudelo, V.; Velásquez-Mejía, E.; Carmona, J.; Abad, J.; Escobar, J. Metformin is associated with higher relative abundance of mucin-degrading akkermansia muciniphila and several short-chain fatty acid-producing microbiota in the gut. Diabetes Care 2017, 40, 54–62. [Google Scholar] [CrossRef] [Green Version]
- Forslund, K.; Hildebrand, F.; Nielsen, T.; Falony, G. Disentangling the effects of type 2 diabetes and metformin on the human gut microbiota. Nature 2016, 528, 262–266. [Google Scholar] [CrossRef] [PubMed]
- Nishijima, S.; Suda, W.; Oshima, K.; Kim, S.W.; Hirose, Y.; Morita, H.; Hattori, M. The gut microbiome of healthy Japanese and its microbial and functional uniqueness. DNA Res. 2016, 23, 125–133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujishiro, M.; Kushiyama, A.; Yamazaki, H.; Kaneko, S.; Koketsu, Y.; Yamamotoya, T.; Kikuchi, T.; Sakoda, H.; Suzuki, R.; Kadowaki, T. Gastrointestinal symptom prevalence depends on disease duration and gastrointestinal region in type 2 diabetes mellitus. World J. Gastroenterol. 2017, 23, 6694–6704. [Google Scholar] [CrossRef] [PubMed]
- Bailey, C. Biguanides and NIDDM. Diabetes Care 1992, 15, 755–772. [Google Scholar] [CrossRef]
- Garber, A.; Duncan, T.; Goodman, A.; Mills, D.; Rohlf, J. Efficacy of Metformin in Type II Diabetes: Results of a Double-Blind, Placebo-Controlled, Dose-Response Trial. Am. J. Med. 1997, 102, 491–497. [Google Scholar] [CrossRef]
- Kistler, B.; Biruete, A.; Chapman-Novakofski, K.; Wilund, K. The Relationship between Intradialytic Nutrition and Gastrointestinal Symptoms Using a Modified Version of the Gastrointestinal Symptom Rating Scale. J. Ren. Nutr. 2018, 28, 129–134. [Google Scholar] [CrossRef]
- Turan, N.; Aşt, T.; Kaya, N. Reliability and Validity of the Turkish Version of the Gastrointestinal Symptom Rating Scale. Gastroenterol. Nurs. 2017, 40, 47–55. [Google Scholar] [CrossRef]
- Souza, G.; Sardá, F.; Giuntini, E.; Gumbrevicius, I.; Morais, M.; Menezes, E. Translation and Validation of the Brazilian Portuguese Version of the Gastrointestinal Symptoms Rating Scale (GSRS) Questionnaire. Arq. Gastroenterol. 2016, 53, 146–151. [Google Scholar] [CrossRef]
- Svedlund, J.; Sjödin, I.; Dotevall, G. GSRS: A clinical rating scale for gastrointestinal symptoms in patients with irritable bowel syndrome and peptic ulcer disease. Dig. Dis. Sci. 1988, 33, 129–134. [Google Scholar] [CrossRef] [PubMed]
- Dimenäs, E.; Glise, H.; Hallerbäck, B.; Hernqvist, H.; Svedlund, J.; Wiklund, I. Quality of life in patients with upper gastrointestinal symptoms. An improved evaluation of treatment regimens? Scand. J. Gastroenterol. 1993, 28, 681–687. [Google Scholar] [CrossRef] [PubMed]
- Adachi, K.; Sugiyama, T.; Yamaguchi, Y.; Tamura, Y.; Izawa, S.; HIjikata, Y.; Ebi, M.; Funai, Y.; Ogasawara, N.; Goto, C.; et al. Gut microbiota disorders cause type 2 diabetes mellitus and homeostatic disturbances in gut-related metabolism in Japanese subjects. J. Clin. Biochem. Nutr. 2019, 64, 231–238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qin, J.; Li, Y.; Cai, Z.; Li, S.; Zhu, J.; Zhang, F.; Liang, S.; Zhang, W.; Guan, Y.; Shen, D.; et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 2012, 490, 55–60. [Google Scholar] [CrossRef]
- Karlsson, F.; Tremaroli, V.; Nookaew, I.; Bergström, G.; Behre, C.; Fagerberg, B.; Nielsen, J.; Bäckhed, F. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature 2013, 498, 99–103. [Google Scholar] [CrossRef]
- Turnbaugh, P.; Ley, R.; Mahowald, M.; Magrini, V.; Mardis, E.; Gordon, J. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006, 444, 1027–1031. [Google Scholar] [CrossRef]
- Patkee, W.; Carr, G.; Baker, E.; Baines, D.; Garnett, J. Metformin prevents the effects of Pseudomonas aeruginosa on airway epithelial tight junctions and restricts hyperglycaemia-induced bacterial growth. J. Cell. Mol. Med. 2016, 20, 758–764. [Google Scholar] [CrossRef] [Green Version]
- Nasrin, F. Study of Antimicrobial and Antioxidant Potentiality of Anti-Diabetic Drug Metformin. Int. J. Pharm. Drug. Anal. 2014, 2, 220–224. [Google Scholar]
- Abbas, H.; Elsherbini, A.; Shaldam, M. Repurposing metformin as a quorum sensing inhibitor in Pseudomonas aeruginosa. Afr. Health Sci. 2017, 17, 808–819. [Google Scholar] [CrossRef] [Green Version]
- Xiao, Y.; Liu, F.; Li, S.; Jiang, N.; Yu, C.; Zhu, X.; Qin, Y.; Hui, J.; Meng, L.; Song, C.; et al. Metformin promotes innate immunity through a conserved PMK-1/p38 MAPK pathway. Virulence 2020, 11, 39–48. [Google Scholar] [CrossRef] [Green Version]
- Lopetuso, L.R.; Petito, V.; Graziani, C.; Schiavoni, E.; Sterbini, F.; Poscia, A.; Scaldaferri, F.; Gaetani, E.; Franceschi, F.; Cammarota, G.; et al. Gut Microbiota in Health, Diverticular Disease, Irritable Bowel Syndrome, and Inflammatory Bowel Diseases: Time for Microbial Marker of Gastrointestinal Disorders. Dig. Dis. 2018, 36, 56–65. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.J.; Lin, C.S.; Lu, C.C.; Martel, J.; Ko, Y.F.; Ojcius, D.; Tseng, S.F.; Wu, T.R.; Chen, Y.Y.; Young, J.; et al. Ganoderma lucidum reduces obesity in mice by modulating the composition of the gut microbiota. Nat. Commun. 2015, 6, 7489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kverka, M.; Zakostelska, Z.; Klimesova, K.; Sokol, D.; Hudcovic, T.; Hrncir, T.; Rossmann, P.; Mrazek, J.; Kopecny, J.; Verdu, E. Oral administration of Parabacteroides distasonis antigens attenuates experimental murine colitis through modulation of immunity and microbiota composition. Clin. Exp. Immunol. 2011, 163, 250–259. [Google Scholar] [CrossRef] [PubMed]
- Shirasawa, Y.; Shibahara-Sone, H.; Iino, T.; Ishikawa, F. Bifidobacterium bifidum BF-1 suppresses Helicobacter pylori-induced genes in human epithelial cells. J. Dairy Sci. 2010, 93, 4526–4534. [Google Scholar] [CrossRef] [Green Version]
- Gomi, A.; Yamaji, K.; Watanabe, O.; Yoshioka, M.; Miyazaki, K.; Iwama, Y.; Urita, Y. Bifidobacterium bifidum YIT 10347 fermented milk exerts beneficial effects on gastrointestinal discomfort and symptoms in healthy adults: A double-blind, randomized, placebo-controlled study. J. Dairy Sci. 2018, 101, 4830–4841. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhuang, X.; Tian, Z.; Li, L.; Zeng, Z.; Chen, M.; Xiong, L. Fecal microbiota alterations associated with diarrhea-predominant irritable bowel syndrome. Front. Microbiol. 2018, 9, 1600. [Google Scholar] [CrossRef] [PubMed]
- Kelly, T.; Bazzano, L.; Ajami, N.; He, H.; Zhao, J.; Petrosino, J.; Correa, A.; He, J. Gut Microbiome Associates with Lifetime Cardiovascular Disease Risk Profile among Bogalusa Heart Study Participants. Circ. Res. 2016, 119, 956–964. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Ajami, N.J.; El-Serag, H.B.; Hair, C.; Graham, D.Y.; White, D.L.; Chen, L.; Wang, Z.; Plew, S.; Ketwaroo, G.; et al. Dietary quality and the colonic mucosa-associated gut microbiome in humans. Am. J. Clin. Nutr. 2019, 110, 701–712. [Google Scholar] [CrossRef]
- Matsuo, S.; Imai, E.; Horio, M.; Yasuda, Y.; Tomita, K.; Nitta, K.; Yamagata, K.; Tomino, Y.; Yokoyama, H.; Hishida, A. Revised equations for estimated GFR from serum creatinine in Japan. Am. J. Kidney Dis. 2009, 53, 982–992. [Google Scholar] [CrossRef]
- Matsuki, T.; Watanabe, K.; Fujimoto, J.; Kado, Y.; Takada, T.; Matsumoto, K.; Tanaka, R. Quantitative PCR with 16S rRNA-Gene-Targeted Species-Specific Primers for Analysis of Human Intestinal Bifidobacteria. Appl. Environ. Microbiol. 2004, 70, 167–173. [Google Scholar] [CrossRef] [Green Version]
- Fadrosh, D.; Ma, B.; Gajer, P.; Sengamalay, N.; Ott, S.; Brotman, R.; Ravel, J. An Improved Dual-Indexing Approach for Multiplexed 16S rRNA Gene Sequencing on the Illumina MiSeq Platform. Microbiome 2014, 2, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caporaso, J.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.; Costello, E.; Fierer, N.; Pẽa, A.G.; Goodrich, J.; Gordon, J.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [Green Version]
- Lozupone, C.; Knight, R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005, 71, 8228–8235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watts, P.; Buleyt, K.; Sandersont, S.; Boardmant, W.; Ciofi, C.; Gibson, R. Human gut microbes associated with obesity. Nature 2006, 444, 1022–1023. [Google Scholar]
- Kadota, K.; Nakai, Y.; Shimizu, K. A weighted average difference method for detecting differentially expressed genes from microarray data. Algorithms Mol. Biol. 2008, 3, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benjamini, Y. Controlling The False Discovery Rate-A Practical and Powerful Approach to Multiple Testing. J. R. Statist. Soc. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liao, X.; Song, L.; Zeng, B.; Liu, B.; Qiu, Y.; Qu, H.; Zheng, Y.; Long, M.; Zhou, H.; Wang, Y. Alteration of gut microbiota induced by DPP-4i treatment improves glucose homeostasis. EBioMedicine 2019, 44, 665–674. [Google Scholar] [CrossRef] [Green Version]
- Naito, Y.; Kashiwagi, K.; Takagi, T.; Andoh, A. Intestinal Dysbiosis Secondary to Proton-Pump Inhibitor Use. Digestion 2018, 97, 195–204. [Google Scholar] [CrossRef]
- Gao, C.; Major, A.; Rendon, D.; Lugo, M.; Jackson, V.; Shi, Z.; Mori-Akiyama, Y.; Versalovic, J. Histamine H2 receptor-mediated suppression of intestinal inflammation by probiotic lactobacillus reuteri. MBio 2015, 6, e01358-15. [Google Scholar] [CrossRef] [Green Version]







| Variables | Total | Subgroup |
|---|---|---|
| n | 31 | 21 |
| Age (years) | 63.3 ± 9.5 | 60.1 ± 9.8 |
| Male | 20 (64.5) | 15 (71.4) |
| Body mass index (kg/m2) | 23.3 ± 3.2 | 24.1 ± 3.0 |
| SBP (mmHg) | 126.2 ± 14.8 | 127.9 ± 14.6 |
| DBP (mmHg) | 71.5 ± 11.7 | 73.4 ± 10.9 |
| PPG (mmol/L) | 7.49 ± 1.63 | 6.95 ± 1.09 |
| Hemoglobin A1c (mmol/mol) | 55.4 ± 7.9 | 53.7 ± 6.1 |
| Hemoglobin A1c (%) | 7.06 ± 0.56 | 7.06 ± 0.56 |
| eGFR (mL/min/1.73 m2) | 79.7 ± 13.7 | 80.1 ± 14.9 |
| Duration of diabetes, (years) | 11.5 ± 11.0 | 8.3 ± 8.3 |
| Diabetic microangiopathy | ||
| Distal symmetric polyneuropathy | 2 (6.5) | 0 (0) |
| Retinopathy | 3 (9.7) | 2 (9.5) |
| Nephropathy | 4 (12.9) | 3 (14.3) |
| H2 blocker | 1 (3.2) | - |
| Proton pump inhibitor | 4 (12.9) | - |
| α-glucosidase inhibitor | 1 (3.2) | - |
| DPP4 inhibitor | 9 (29.0) | - |
| Total | Baseline | After 2 Weeks | After 4 Weeks | p-Value |
|---|---|---|---|---|
| Hemoglobin A1c (mmol/mol) | 55.4 ± 7.9 | - | 52.5 ± 6.6 | 0.0003 |
| Hemoglobin A1c (%) | 7.2 ± 0.7 | - | 7.0 ± 0.6 | |
| Total score | 20.3 ± 5.8 | 21.0 ± 5.4 | 22.8 ± 7.1 | 0.04 |
| Subscale | ||||
| Abdominal pain | 3.6 ± 1.2 | 3.9 ± 1.7 | 3.9 ± 1.6 | 0.24 |
| Regurgitation | 2.7 ± 1.6 | 2.8 ± 1.5 | 2.8 ± 1.7 | 0.69 |
| Dyspepsia | 6.0 ± 2.1 | 6.0 ± 1.9 | 6.8 ± 2.7 | 0.04 |
| Diarrhea | 2.9 ± 1.3 | 3.2 ± 1.4 | 3.3 ± 1.7 | 0.34 |
| Constipation | 5.0 ± 2.2 | 5.5 ± 2.5 | 5.9 ± 2.2 | 0.05 |
| Bristol Stool Form Scale | 3.7 ± 0.9 | 3.9 ± 1.0 | 3.9 ± 1.4 | 0.44 |
| Subgroup | Baseline | After 2 Weeks | After 4 Weeks | p-Value |
| Hemoglobin A1c (mmol/mol) | 53.7 ± 6.1 | - | 52.3 ± 6.4 | 0.0005 |
| Hemoglobin A1c (%) | 7.1 ± 0.6 | - | 6.9 ± 0.6 | |
| Total score | 20.0 ± 6.0 | 20.3 ± 4.6 | 22.6 ± 7.7 | 0.12 |
| Subscale | ||||
| Abdominal pain | 3.5 ± 1.1 | 3.7 ± 1.3 | 3.8 ± 1.7 | 0.34 |
| Regurgitation | 2.7 ± 1.7 | 2.8 ± 1.6 | 2.9 ± 2.0 | 0.45 |
| Dyspepsia | 5.9 ± 2.0 | 5.9 ± 1.9 | 6.7 ± 2.9 | 0.16 |
| Diarrhea | 3.1 ± 1.4 | 3.2 ± 1.4 | 3.3 ± 1.7 | 0.83 |
| Constipation | 4.7 ± 1.9 | 5.2 ± 2.1 | 5.9 ± 2.3 | 0.03 |
| Bristol Stool Form Scale | 3.7 ± 0.9 | 3.9 ± 1.0 | 3.9 ± 1.4 | 0.12 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nakajima, H.; Takewaki, F.; Hashimoto, Y.; Kajiyama, S.; Majima, S.; Okada, H.; Senmaru, T.; Ushigome, E.; Nakanishi, N.; Hamaguchi, M.; et al. The Effects of Metformin on the Gut Microbiota of Patients with Type 2 Diabetes: A Two-Center, Quasi-Experimental Study. Life 2020, 10, 195. https://0-doi-org.brum.beds.ac.uk/10.3390/life10090195
Nakajima H, Takewaki F, Hashimoto Y, Kajiyama S, Majima S, Okada H, Senmaru T, Ushigome E, Nakanishi N, Hamaguchi M, et al. The Effects of Metformin on the Gut Microbiota of Patients with Type 2 Diabetes: A Two-Center, Quasi-Experimental Study. Life. 2020; 10(9):195. https://0-doi-org.brum.beds.ac.uk/10.3390/life10090195
Chicago/Turabian StyleNakajima, Hanako, Fumie Takewaki, Yoshitaka Hashimoto, Shizuo Kajiyama, Saori Majima, Hiroshi Okada, Takafumi Senmaru, Emi Ushigome, Naoko Nakanishi, Masahide Hamaguchi, and et al. 2020. "The Effects of Metformin on the Gut Microbiota of Patients with Type 2 Diabetes: A Two-Center, Quasi-Experimental Study" Life 10, no. 9: 195. https://0-doi-org.brum.beds.ac.uk/10.3390/life10090195