Counterion Effect and Isostructurality in a Series of Ag(I) Complexes Containing a Flexible, Imidazole Based Dipodal Ligand
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents and Materials
2.2. Structure Determination
3. Results
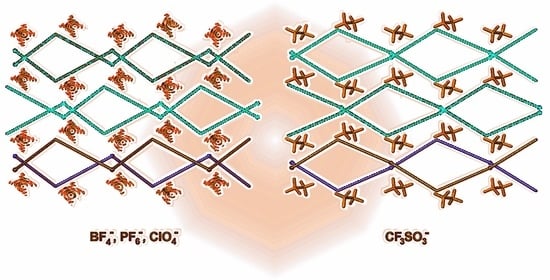
Crystal Structures of Complexes 1–4: {[AgL]counterion}n
4. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- IUCr. Available online: http://reference.iucr.org/dictionary/Isostructural_crystals (accessed on 13 February 2021).
- Mondal, P.K.; Shukla, R.; Biswas, S.; Chopra, D. Role of halogen-involved intermolecular interactions and existence of isostructurality in the crystal packing of—CF3 and halogen (Cl or Br or I) substituted benzamides. Acta Crystallogr. 2018, B74, 1–18. [Google Scholar] [CrossRef]
- Dey, D.; Chopra, D. Occurrence of 3D isostructurality in fluorinated phenyl benzamidines. CrystEngComm 2017, 19, 47–63. [Google Scholar] [CrossRef] [Green Version]
- Mazur, L.; Koziol, A.E.; Jarzembska, K.N.; Paprocka, R.; Modzelewska-Banachiewicz, B. Polymorphism and isostructurality of the series of 3-(4,5-diaryl-4H-1,2,4-triazole-3-yl)propenoic acid derivatives. Cryst. Growth Des. 2017, 17, 2104–2115. [Google Scholar] [CrossRef]
- Suresh, K.; Khandavilli, U.B.R.; Gunnam, A.; Nangia, A. Polymorphism, isostructurality and physicochemical properties of glibenclamide salts. CrystEngComm 2017, 19, 918–929. [Google Scholar] [CrossRef]
- Sridhar, B.; Nanubolu, J.B.; Ravikumar, K.; Karthik, B.; Reddy, B.V.S. Three isostructural solvates of a tetrahydrofurochromenone derivative. Acta Crystallogr. 2017, C73, 407–413. [Google Scholar] [CrossRef]
- Czylkowska, A.; Pietrzak, A.; Szczesio, M.; Rogalewicz, B.; Wojciechowski, J. Crystal structures, Hirshfeld surfaces, and thermal study of isostructural polymeric ladders of La(III) and Sm(III) coordination compounds with 4,4’-Bipyridine and Dibromoacetates. Materials 2020, 13, 4274. [Google Scholar] [CrossRef] [PubMed]
- Kálmán, A.; Párkányi, L.; Argay, G. Classification of the Isostructurality of Organic Molecules in the Crystalline State. Acta Crystallogr. 1993, B49, 1039–1049. [Google Scholar] [CrossRef]
- Fábián, L.; Kálmán, A. Volumetric measure of isostructurality. Acta Crystallogr. 1999, B55, 1099–1108. [Google Scholar] [CrossRef]
- Kálmán, A.; Párkányi, L. Isostructurality of Organic Crystals in Advances in Molecular Structure Research; Hargittai, M., Hargittai, I., Eds.; JAI Press Inc: Greenwich, CT, USA, 1997; Volume 3, pp. 189–226. [Google Scholar]
- Kálmán, A. In Fundamental Principles of Molecular Modeling; Gans, W., Ed.; Plenum Press: New York, NY, USA, 1996; p. 209. [Google Scholar]
- Fábián, L.; Kálmán, A. Isostructurality in one and two dimensions: Isostructurality of polymorphs. Acta Crystallogr. 2004, B60, 547–558. [Google Scholar] [CrossRef]
- Coles, S.J.; Threlfall, T.L.; Tizzard, G.J. The Same but Different: Isostructural Polymorphs and the Case of 3-Chloromandelic Acid. Cryst. Growth Des. 2014, 14, 1623–1628. [Google Scholar] [CrossRef]
- Deya, D.; Thomas, S.P.; Spackman, M.A.; Chopra, D. Quasi-isostructural polymorphism in molecular crystals:Inputs from interaction hierarchy and energyframeworks. Chem. Commun. 2016, 52, 2141–2144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jha, K.K.; Dutta, S.; Kumar, V.; Munshi, P. Isostructural Polymorphs: Qualitative Insights from Energy Frameworks. CrystEngComm 2016, 18, 8497–8505. [Google Scholar] [CrossRef]
- Gelbrich, T.; Hursthouse, M.B. A versatile procedure for the identification, description and quantification of structural similarity in molecular crystals. CrystEngComm 2005, 7, 324–336. [Google Scholar] [CrossRef]
- Alen, J.; Van Meervelt, L.; Dehaen, W.; Dobrzańska, L. Solvent diffusion through a non-porous crystal ‘caught in the act’ and related single-crystal-to-single-crystal transformations in a cationic dinuclear Ag(I) complex. CrystEngComm 2015, 17, 8957–8964. [Google Scholar] [CrossRef]
- Dobrzańska, L. Concomitant, genuine 1D supramolecular isomers of an Ag(I) complex with 1,3-bis(imidazol-1-ylmethyl)-2,4,6-trimethylbenzene and BF4− as counterion. Inorg. Chem. Commun. 2015, 55, 21–24. [Google Scholar] [CrossRef]
- Dobrzańska, L. Anion directed supramolecular architectures of silver(I) complexes with 1,3-bis(imidazol-1-ylmethyl)-2,4,6- trimethylbenzene and a reversible, solvent-induced structural change during a single-crystal-to-single-crystal transformation. CrystEngComm 2011, 13, 2303–2309. [Google Scholar] [CrossRef]
- Arhangelskis, M.; Van Meervelt, L.; Dobrzańska, L. Influence of ligand composition on crystal structure formation—Isostructurality and morphotropism. CrystEngComm 2021, 23, 317–323. [Google Scholar] [CrossRef]
- Sui, B.; Fana, J.; Okamura, T.; Sun, W.-Y.; Ueyama, N. Synthesis, structure and properties of Mn(II), Zn(II), Ag(I) and Cu(II) complexes with 1,3-bis(imidazole-1-ylmethyl)-5-methylbenzene. Solid State Sci. 2005, 7, 969–982, structure herein used to determine certain crystallographic values—no esd values available. [Google Scholar] [CrossRef]
- Ma, Y.; Huang, W.; Yao, J.; Li, B.; Gou, S.; Fun, H.-K. A three-dimensional zinc(II) interpenetrating network and a π–π induced silver(I) zigzag complex connected by 1,3-di(imidazole-1-yl-methyl)-5-methylbenzene. J. Mol. Str. 2003, 658, 51–58. [Google Scholar] [CrossRef]
- Rigaku Oxford Diffraction. CrysAlisPro Software System, version 1.171.39.46; Rigaku Corporation: Oxford, UK, 2018. [Google Scholar]
- Sheldrick, G.M. A short history of SHELX. Acta Crystallogr. 2008, A64, 112–122. [Google Scholar] [CrossRef] [Green Version]
- Sheldrick, G.M. Crystal structure refinement with SHELXL. Acta Crystallogr. 2015, C71, 3–8. [Google Scholar]
- Macrae, C.F.; Bruno, I.J.; Chisholm, J.A.; Edgington, P.R.; McCabe, P.; Pidcock, E.; Rodriguez-Monge, L.; Taylor, R.; van de Streek, J.; Wood, P.A. New Features for the Visualization and Investigation of Crystal Structures. J. Appl. Crystallogr. 2008, 41, 466–470. [Google Scholar] [CrossRef]
- Available online: www.povray.org.
- Fan, J.; Sun, W.-Y.; Okamura, T.; Zheng, Y.-Q.; Sui, B.; Tang, W.-X.; Ueyama, N. Novel Metal-Organic Frameworks with Specific Topology Formed through Noncovalent Br···Br Interactions in the Solid State. Cryst. Growth Des. 2004, 4, 579–584. [Google Scholar] [CrossRef]
- Arunan, E.; Desiraju, G.R.; Klein, R.A.; Sadlej, J.; Scheiner, S.; Alkorta, I.; Clary, D.C.; Crabtree, R.H.; Dannenberg, J.J.; Hobza, P.; et al. Definition of the hydrogen bond (IUPAC Recommendations 2011). Pure Appl. Chem. 2011, 83, 1637–1641. [Google Scholar] [CrossRef]
- Arunan, E.; Desiraju, G.R.; Klein, R.A.; Sadlej, J.; Scheiner, S.; Alkorta, I.; Clary, D.C.; Crabtree, R.H.; Dannenberg, J.J.; Hobza, P.; et al. Defining the hydrogen bond: An account (IUPAC Technical Report). Pure Appl. Chem. 2011, 83, 1619–1636. [Google Scholar] [CrossRef]
- Desiraju, G.R.; Steiner, T. The Weak Hydrogen Bond in Structural Chemistry and Biology; Oxford University Press: Oxford, UK, 2006; p. 13. [Google Scholar]
- Wolff, S.K.; Grimwood, D.J.; McKinnon, J.J.; Turner, M.J.; Jayatilaka, D.; Spackman, M.A. CrystalExplorer (Version 3.1); University of Western Australia: Perth, Australia, 2012. [Google Scholar]
- Spackman, M.A.; McKinnon, J.J. Fingerprinting intermolecular interactions in molecular crystals. CrystEngComm 2002, 4, 378–392. [Google Scholar] [CrossRef]
- Mingos, D.M.P.; Rohl, A.L. Size and Shape Characteristics of Inorganic Molecules and Ions and their Relevance to Molecular Packing Problems. J. Chem. Soc. Dalton Trans. 1991, 12, 3419–3425. [Google Scholar] [CrossRef]
- Wood, P.A.; Oliveira, M.A.; Zink, A.; Hickey, M.B. Isostructurality in pharmaceutical salts: How often and how similar? CrystEngComm 2012, 14, 2413–2421. [Google Scholar] [CrossRef]
- Georgiadou, D.G.; Palilis, L.C.; Vasilopoulou, M.; Pistolis, G.; Dimotikali, D.; Argitis, P. Influence of the anion on the optoelectronic characteristics of triphenylsulfonium salts modified polymer light emitting devices. Synth. Met. 2013, 181, 37–44. [Google Scholar] [CrossRef]
- Spek, A.L. Structure validation in chemical crystallography. Acta Crystallogr. 2009, D65, 148–155. [Google Scholar] [CrossRef]
- Phuengphai, P.; Massera, C.; Reedijk, J.; Youngme, S.; Gamez, P. Anion Exchange in Coordination-Network Materials. Eur. J. Inorg. Chem. 2013, 27, 4812–4822. [Google Scholar] [CrossRef]







| Parameter | Compound Reference | |
|---|---|---|
| 1 | 2 | |
| Chemical formula | C15H16AgN4·BF4 | C15H16AgN4·PF6 |
| Formula Mass | 447.00 | 505.16 |
| Crystal system | Monoclinic | Monoclinic |
| a/Å | 13.3836(7) | 13.2476(13) |
| b/Å | 14.9566(8) | 15.5277(10) |
| c/Å | 8.3713(5) | 8.8493(10) |
| α/° | 90 | 90 |
| β/° | 106.836(6) | 105.538(9) |
| γ/° | 90 | 90 |
| Unit cell volume/Å3 | 1603.88(16) | 1753.8(3) |
| Temperature/K | 100(2) | 100(2) |
| Space group | Cc | Cc |
| No. of formula units per unit cell, Z | 4 | 4 |
| Radiation type | Mo Kα | Mo Kα |
| Absorption coefficient, μ/mm−1 | 1.305 | 1.309 |
| No. of reflections measured | 11179 | 3791 |
| No. of independent reflections | 3787 | 2557 |
| Rint | 0.0313 | 0.0252 |
| Final R1 a values (I > 2σ(I)) | 0.0252 | 0.0378 |
| Final wR2 b values (I > 2σ(I)) | 0.0541 | 0.0773 |
| Final R1 a values (all data) | 0.0273 | 0.0492 |
| Final wR2 b values (all data) | 0.0554 | 0.0820 |
| Goodness of fit on F2 Flack parameter CCDC number | 1.053 −0.02(1) 2065080 | 1.033 −0.05(3) 2065081 |
| Compound | D-H···A | H···A/Å | D···A/Å | D-H···A/° |
|---|---|---|---|---|
| 1 | C8-H8···Cgi (imidazole: 1–5) | 2.98 | 3.887(5) | 160 |
| C14-H14B···Cgii (benzene) | 2.82 | 3.449(5) | 122 | |
| C12-H12···F21 | 2.47 | 3.420(4) | 176 | |
| C18-H18···F21iii | 2.42 | 3.324(5) | 159 | |
| C2-H2···F22iv | 2.42 | 3.142(5) | 132 | |
| C6-H6B···F22v | 2.51 | 3.306(5) | 161 | |
| C13-H13B···F23iv | 2.72 | 3.554(6) | 143 | |
| C19-H19···F23vi | 2.87 | 3.754(6) | 156 | |
| C6-H6A···F24 | 2.48 | 3.442(6) | 164 | |
| C13-H13C···F24ii | 2.38 | 3.327(5) | 161 | |
| C10-H10···F24vi | 2.76 | 3.493(6) | 135 | |
| 2 | C14-H14B···Cgi (benzene) | 2.96 | 3.507(10) | 116 |
| C2-H2···F21ii | 2.81 | 3.742(10) | 165 | |
| C13-H13B···F21ii | 2.78 | 3.665(12) | 151 | |
| C10-H10···F23i | 2.37 | 3.194(11) | 145 | |
| C14-H14B···F23iii | 2.90 | 3.627(11) | 131 | |
| C19-H19···F23iii | 2.83 | 3.608(11) | 141 | |
| C12-H12···F24 | 2.51 | 3.462(8) | 177 | |
| C13-H13A···F24i | 2.75 | 3.709(11) | 167 | |
| C18-H18···F24iv | 2.71 | 3.653(10) | 170 | |
| C13-H13C···F25iii | 2.99 | 3.896(11) | 154 | |
| C6-H6B···F26v | 2.86 | 3.485(11) | 122 | |
| 3 | C11-H5···Cgi (benzene) | 2.89 | 3.499(8) | 120 |
| C12-H6···O1ii | 2.76 | 3.568(10) | 143 | |
| C51-H15···O1iii | 2.72 | 3.588(11) | 148 | |
| C14-H8···O1iv | 2.90 | 3.785(10) | 156 | |
| C2-H1···O2ii | 2.56 | 3.506(10) | 178 | |
| C13-H7···O2v | 2.49 | 3.410(10) | 164 | |
| C51---H14···O2vi | 2.86 | 3.837(11) | 174 | |
| C31-H9···O3ii | 2.54 | 3.512(9) | 167 | |
| C6-H3···O3iv | 2.88 | 3.590(10) | 132 | |
| C51-H16···O3iv | 2.46 | 3.396(10) | 159 | |
| C32-H11···O4iii | 2.47 | 3.200(9) | 134 | |
| C31-H10···O4vii | 2.63 | 3.408(10) | 135 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dobrzańska, L. Counterion Effect and Isostructurality in a Series of Ag(I) Complexes Containing a Flexible, Imidazole Based Dipodal Ligand. Materials 2021, 14, 1804. https://0-doi-org.brum.beds.ac.uk/10.3390/ma14071804
Dobrzańska L. Counterion Effect and Isostructurality in a Series of Ag(I) Complexes Containing a Flexible, Imidazole Based Dipodal Ligand. Materials. 2021; 14(7):1804. https://0-doi-org.brum.beds.ac.uk/10.3390/ma14071804
Chicago/Turabian StyleDobrzańska, Liliana. 2021. "Counterion Effect and Isostructurality in a Series of Ag(I) Complexes Containing a Flexible, Imidazole Based Dipodal Ligand" Materials 14, no. 7: 1804. https://0-doi-org.brum.beds.ac.uk/10.3390/ma14071804