Enzymatic Hydrolysis of Alginate to Produce Oligosaccharides by a New Purified Endo-Type Alginate Lyase
Abstract
:1. Introduction
2. Results and Discussion
2.1. Isolation of Alginate-Degrading Strain
2.2. Purification of Alginate Lyase
2.3. Substrate Specifities and Enzymatic Kinetics of the Enzyme
2.4. Biochemical Properties of the Enzyme
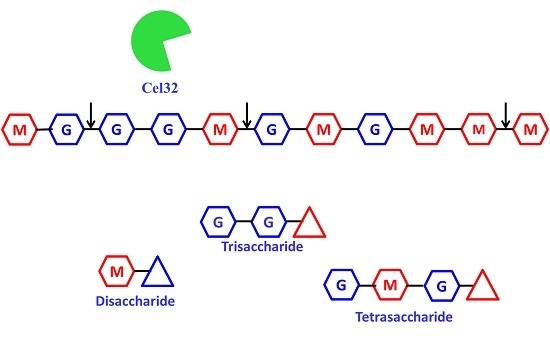
2.5. The Action Modes and Substrate Binding Subsites of the Enzyme
2.6. Enzymatic Hydrolysis of Sodiumalginate by Using Cel32
2.7. ESI-MS Analysis of the Degradation Products of Cel32
3. Materials and Methods
3.1. Microorganism
3.2. Media and Culture Conditions
3.3. Strain Isolation and Identification
3.4. Enzyme Purification
3.5. Assay for Alginate Lyase Activity and Protein Concentration
3.6. Substrate Specificity and Kinetic Measurement of Alginate Lyase
3.7. Biochemical Characterization of the Enzyme
3.8. Analysis of Action Mode and Substrate Binding of the Enzyme
3.9. Enzymatic Depolymerization of Sodium Alginate
3.10. ESI-MS Analysis of the Degradation Products of the Enzyme
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Gacesa, P. Enzymic degradation of alginates. Int. J. Biochem. 1992, 24, 545–552. [Google Scholar] [CrossRef]
- Pawar, S.N.; Edgar, K.J. Alginate derivatization: A review of chemistry, properties and applications. Biomaterials 2012, 33, 3279–3305. [Google Scholar] [CrossRef] [PubMed]
- Fujihara, M.; Nagumo, T. An influence of the structure of alginate on the chemotactic activity of macrophages and the antitumor activity. Carbohydr. Res. 1993, 243, 211–216. [Google Scholar] [CrossRef]
- Otterlei, M.; Ostgaard, K.; Skjak-Braek, G.; Smidsrod, O.; Soon-Shiong, P.; Espevik, T. Induction of cytokine production from human monocytes stimulated with alginate. J. Immunother. 1991, 10, 286–291. [Google Scholar] [CrossRef] [PubMed]
- Tusi, S.K.; Khalaj, L.; Ashabi, G.; Kiaei, M.; Khodagholi, F. Alginate oligosaccharide protects against endoplasmic reticulum- and mitochondrial-mediated apoptotic cell death and oxidative stress. Biomaterials 2011, 32, 5438–5458. [Google Scholar] [CrossRef] [PubMed]
- Choi, I.; Jo, G.; Kim, S.; Jung, C.W.; Kim, Y.; Shin, K. Stimulation of various functions in murine peritoneal macrophages by glucans produced by glucosyltransferases from Streptococcus mutans. Biosci. Biotechnol. Biochem. 2005, 69, 1693–1699. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.K.; Jiang, X.L.; Hwang, H.M.; Liu, S.L.; Guan, H.S. Promotive effects of alginate-derived oligosaccharide on maize seed germination. J. Appl. Phycol. 2004, 16, 73–76. [Google Scholar] [CrossRef]
- Ueyama, Y.; Ishikawa, K.; Mano, T.; Koyama, T.; Nagatsuka, H.; Suzuki, K.; Ryoke, K. Usefulness as guided bone regeneration membrane of the alginate membrane. Biomaterials 2002, 23, 2027–2033. [Google Scholar] [CrossRef]
- Li, Y.C.; Wang, Z.; Xu, X.D.; Jin, L.Q. A ca-alginate particle co-immobilized with phanerochaete chrysosporium cells and the combined cross-linked enzyme aggregates from Trametes versicolor. Bioresour. Technol. 2015, 198, 464–469. [Google Scholar] [CrossRef] [PubMed]
- Iwamoto, M.; Kurachi, M.; Nakashima, T.; Kim, D.; Yamaguchi, K.; Oda, T.; Iwamoto, Y.; Muramatsu, T. Structure-activity relationship of alginate oligosaccharides in the induction of cytokine production from RAW264.7 cells. FEBS Lett. 2005, 579, 4423–4429. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.X.; Zhou, J.H.; Ha, Z.P.; Zhang, Y.X.; Gu, G.M. Hypoglycemic effect of Rehmannia glutinosa oligosaccharide in hyperglycemic and alloxan-induced diabetic rats and its mechanism. J. Ethnopharmacol. 2004, 90, 39–43. [Google Scholar] [CrossRef] [PubMed]
- Wong, T.Y.; Preston, L.A.; Schiller, N.L. Alginate lyase: Review of major sources and enzyme characteristics, structure-function analysis, biological roles, and applications. Annu. Rev. Microbiol. 2000, 54, 289–340. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.W.; Tan, H.D.; Qin, Y.Q.; Xu, Q.S.; Du, Y.G.; Yin, H. Characterization of a new endo-type alginate lyase from Vibrio sp. W13. Int. J. Biol. Macromol. 2015, 75, 330–337. [Google Scholar] [CrossRef] [PubMed]
- Dou, W.F.; Wei, D.; Li, H.; Li, H.; Rahman, M.M.; Shi, J.S.; Xu, Z.H.; Ma, Y.H. Purification and characterisation of a bifunctional alginate lyase from novel Isoptericola halotolerans CGMCC 5336. Carbohyd. Polym. 2013, 98, 1476–1482. [Google Scholar] [CrossRef] [PubMed]
- Li, J.W.; Dong, S.; Song, J.; Li, C.B.; Chen, X.L.; Xie, B.B.; Zhang, Y.Z. Purification and characterization of a bifunctional alginate lyase from Pseudoalteromonas sp. SM0524. Mar. Drugs 2011, 9, 109–123. [Google Scholar] [CrossRef] [PubMed]
- Iwamoto, Y.; Araki, R.; Iriyama, K.; Oda, T.; Fukuda, H.; Hayashida, S.; Muramatsu, T. Purification and characterization of bifunctional alginate lyase from Alteromonas sp. strain No. 272 and its action on saturated oligomeric substrates. Biosci. Biotechnol. Biochem. 2001, 65, 133–142. [Google Scholar] [CrossRef] [PubMed]
- Matsushima, R.; Danno, H.; Uchida, M.; Ishihara, K.; Suzuki, T.; Kaneniwa, M.; Ohtsubo, Y.; Nagata, Y.; Tsuda, M. Analysis of extracellular alginate lyase and its gene from a marine bacterial strain, Pseudoalteromonas atlantica AR06. Appl. Microbiol. Biotechnol. 2010, 86, 567–576. [Google Scholar] [CrossRef] [PubMed]
- Sawabe, T.; Takahashi, H.; Ezura, Y.; Gacesa, P. Cloning, sequence analysis and expression of Pseudoalteromonas elyakovii IAM 14594 gene (alyPEEC) encoding the extracellular alginate lyase. Carbohyd. Res. 2001, 335, 11–21. [Google Scholar] [CrossRef]
- Badur, A.H.; Jagtap, S.S.; Yalamanchili, G.; Lee, J.K.; Zhao, H.; Rao, C.V. Alginate lyases from alginate-degrading Vibrio splendidus 12B01 are endolytic. Appl. Environ. Microbiol. 2015, 81, 1865–1873. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.T.; Chung, J.H.; Wang, D.; Lee, J.; Woo, H.C.; Choi, I.G.; Kim, K.H. Depolymerization of alginate into a monomeric sugar acid using Alg17C, an exo-oligoalginate lyase cloned from Saccharophagus degradans 2-40. Appl. Microbiol. Biotechnol. 2012, 93, 2233–2239. [Google Scholar] [CrossRef] [PubMed]
- Park, H.H.; Kam, N.; Lee, E.Y.; Kim, H.S. Cloning and characterization of a novel oligoalginate lyase from a newly isolated bacterium Sphingomonas sp. MJ-3. Mar. Biotechnol. 2012, 14, 189–202. [Google Scholar] [CrossRef] [PubMed]
- Jagtap, S.S.; Hehemann, J.H.; Polz, M.F.; Lee, J.K.; Zhao, H.M. Comparative biochemical characterization of three exolytic oligoalginate lyases from Vibrio splendidus reveals complementary substrate scope, temperature, and pH adaptations. Appl. Environ. Microbiol. 2014, 80, 4207–4214. [Google Scholar] [CrossRef] [PubMed]
- Li, L.Y.; Jiang, X.L.; Guan, H.S.; Wang, P. Preparation, purification and characterization of alginate oligosaccharides degraded by alginate lyase from Pseudomonas sp. HZJ 216. Carbohyd. Res. 2011, 346, 794–800. [Google Scholar] [CrossRef] [PubMed]
- Aarstad, O.A.; Tondervik, A.; Sletta, H.; Skjak-Braek, G. Alginate sequencing: An analysis of block distribution in alginates using specific alginate degrading enzymes. Biomacromolecules 2012, 13, 106–116. [Google Scholar] [CrossRef] [PubMed]
- Inoue, A.; Mashino, C.; Kodama, T.; Ojima, T. Protoplast preparation from Laminaria japonica with recombinant alginate lyase and cellulase. Mar. Biotechnol. 2011, 13, 256–263. [Google Scholar] [CrossRef] [PubMed]
- Xiaoke, H.; Xiaolu, J.; Huashi, G. Isolation of protoplasts from undaria pinnatifida by alginate lyase digestion. J. Ocean Univ. Qingdao 2003, 2, 58–61. [Google Scholar]
- Islan, G.A.; Martinez, Y.N.; Illanes, A.; Castro, G.R. Development of novel alginate lyase cross-linked aggregates for the oral treatment of cystic fibrosis. RSC Adv. 2014, 4, 11758–11765. [Google Scholar] [CrossRef]
- Kobayashi, T.; Uchimura, K.; Miyazaki, M.; Nogi, Y.; Horikoshi, K. A new high-alkaline alginate lyase from a deep-sea bacterium Agarivorans sp. Extremophiles 2009, 13, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.T.; Lin, H.; Kim, S.M. Purification and characterization of a Na+/K+ dependent alginate lyase from turban shell gut Vibrio sp. YKW-34. Enzym. Microb. Technol. 2007, 41, 828–834. [Google Scholar] [CrossRef]
- Inoue, A.; Takadono, K.; Nishiyama, R.; Tajima, K.; Kobayashi, T.; Ojima, T. Characterization of an alginate lyase, FlAlYa, from Flavobacterium sp. strain UMI-01 and its expression in Escherichia coli. Mar. Drugs 2014, 12, 4693–4712. [Google Scholar] [CrossRef] [PubMed]
- Kumar, C.G.; Takagi, H. Microbial alkaline proteases: From a bioindustrial viewpoint. Biotechnol. Adv. 1999, 17, 561–594. [Google Scholar] [CrossRef]
- Sim, S.J.; Baik, K.S.; Park, S.C.; Choe, H.N.; Seong, C.N.; Shin, T.S.; Woo, H.C.; Cho, J.Y.; Kim, D. Characterization of alginate lyase gene using a metagenomic library constructed from the gut microflora of abalone. J. Ind. Microbiol. Biotechnol. 2012, 39, 585–593. [Google Scholar] [CrossRef] [PubMed]
- El-Katatny, M.H.; Hetta, A.M.; Shaban, G.M.; El-Komy, H.M. Improvement of cell wall degrading enzymes production by alginate encapsulated Trichoderma spp. Food Technol. Biotechnol. 2003, 41, 219–225. [Google Scholar]
- Bonjoch, N.; Tamayo, P. Protein content quantification by Bradford method. In Handbook of Plant Ecophysiology Techniques; Reigosa Roger, M., Ed.; Springer: Dordrecht, The Netherlands, 2001; pp. 283–295. [Google Scholar]
- Swift, S.M.; Hudgens, J.W.; Heselpoth, R.D.; Bales, P.M.; Nelson, D.C. Characterization of AlgMsp, an alginate lyase from Microbulbifer sp. 6532A. PLoS ONE 2014, 9, e112939. [Google Scholar] [CrossRef] [PubMed]







| Step | Total Protein (mg) | Total Activity (U) | Specific Activity * (U/mg) | Folds | Recovery (%) |
|---|---|---|---|---|---|
| Crude enzyme | 1331 | 100,548 | 76 | 1 | 100 |
| (NH4)2SO4 precipitation | 73.2 | 37,625 | 514 | 6.8 | 37.4 |
| SephadexG-200-filtration | 0.73 | 9577 | 13,042 | 172.6 | 9.5 |
| SephadexG-75-filtration | 0.28 | 6770 | 24,083 | 318.1 | 6.7 |
| Substrate | Sodium Alginate | PolyMG | PolyM | PolyG |
|---|---|---|---|---|
| Specific activity * (U/mg) | 24,083.21 | 15,952.55 | 30,665.69 | 15,970.07 |
| Km (mM) | 27.21 | 23.90 | 53.61 | 15.62 |
| Vmax (nmol/s) | 4.37 | 3.50 | 7.02 | 2.22 |
| Kcat (s−1) | 101.87 | 81.64 | 163.73 | 51.78 |
| Kcat/Km (s−1/mM) | 3.74 | 3.42 | 3.05 | 3.31 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, B.; Chen, M.; Yin, H.; Du, Y.; Ning, L. Enzymatic Hydrolysis of Alginate to Produce Oligosaccharides by a New Purified Endo-Type Alginate Lyase. Mar. Drugs 2016, 14, 108. https://0-doi-org.brum.beds.ac.uk/10.3390/md14060108
Zhu B, Chen M, Yin H, Du Y, Ning L. Enzymatic Hydrolysis of Alginate to Produce Oligosaccharides by a New Purified Endo-Type Alginate Lyase. Marine Drugs. 2016; 14(6):108. https://0-doi-org.brum.beds.ac.uk/10.3390/md14060108
Chicago/Turabian StyleZhu, Benwei, Meijuan Chen, Heng Yin, Yuguang Du, and Limin Ning. 2016. "Enzymatic Hydrolysis of Alginate to Produce Oligosaccharides by a New Purified Endo-Type Alginate Lyase" Marine Drugs 14, no. 6: 108. https://0-doi-org.brum.beds.ac.uk/10.3390/md14060108