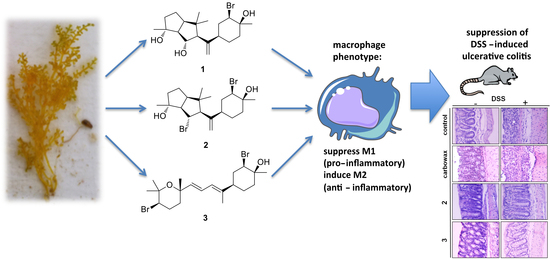
Neorogioltriol and Related Diterpenes from the Red Alga Laurencia Inhibit Inflammatory Bowel Disease in Mice by Suppressing M1 and Promoting M2-Like Macrophage Responses
Abstract
:1. Introduction
2. Results and Discussion
2.1. Isolation of Metabolites 1–3 and Evaluation of their Anti-Inflammatory Capacity
2.2. Macrophages Exposed to Metabolites 1–3 Acquire an M2-Like Anti-Inflammatory Phenotype
2.3. Metabolites 1–3 Suppress M1 Activation of Macrophages but do not Affect Induction of Endotoxin Tolerance
2.4. Evaluating the Effect of Metabolites 2 and 3 in DSS-Induced Colitis in Mice
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Collection of Algal Material
3.3. Isolation of Metabolites 1–3
3.4. Cell Culture
3.5. Cell Proliferation Assay
3.6. Measurement of Nitrite Production
3.7. cDNA Synthesis and Quantitative PCR for Cell Culture Samples
3.8. Elisa
3.9. Mice
3.10. Tissue Processing and Histological Evaluations
3.11. Determination of Biochemical Markers
3.12. Real-Time PCR from Mouse Tissue Samples
3.13. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Daskalaki, M.G.; Tsatsanis, C.; Kampranis, S.C. Histone methylation and acetylation in macrophages as a mechanism for regulation of inflammatory responses. J. Cell Physiol. 2018, 233, 6495–6507. [Google Scholar] [CrossRef] [PubMed]
- Vergadi, E.; Ieronymaki, E.; Lyroni, K.; Vaporidi, K.; Tsatsanis, C. Akt Signaling Pathway in Macrophage activation and M1/M2 Polarization. J. Immunol. 2017, 198, 1006–1014. [Google Scholar] [CrossRef] [PubMed]
- Murray, P.J.; Allen, J.E.; Biswas, S.K.; Fisher, E.A.; Gilroy, D.W.; Goerdt, S.; Gordon, S.; Hamilton, J.A.; Ivashkiv, L.B.; Lawrence, T.; et al. Macrophage activation and polarization: Nomenclature and experimental guidelines. Immunity 2014, 41, 14–20. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, T.; Natoli, G. Transcriptional regulation of macrophage polarization: Enabling diversity with identity. Nat. Rev. Immunol. 2011, 11, 750–761. [Google Scholar] [CrossRef] [PubMed]
- Chatter, R.; Kladi, M.; Tarhouni, S.; Maatoug, R.; Kharrat, R.; Vagias, C.; Roussis, V. Neorogioltriol: A brominated diterpene with analgesic activity from Laurencia glandulifera. Phytochem. Lett. 2009, 2, 25–28. [Google Scholar] [CrossRef]
- Chatter, R.; Othman, R.B.; Rabhi, S.; Kladi, M.; Tarhouni, S.; Vagias, C.; Roussis, V.; Guizani-Tabbane, L.; Kharrat, R. In Vivo and in Vitro Anti-Inflammatory Activity of Neorogioltriol, a New Diterpene Extracted from the Red Algae Laurencia glandulifera. Mar. Drugs 2011, 9, 1293–1306. [Google Scholar] [CrossRef] [PubMed]
- Guella, G.; Pietra, F. A new-skeleton diterpenoid, new prenylbisabolanes, and their putative biogenetic precursor, from the red seaweed Laurencia microcladia from Il Rogiolo: Assigning the absolute configuration when two chiral halves are connected by single bonds. Helv. Chim. Acta 2000, 83, 2946–2952. [Google Scholar] [CrossRef]
- Kokkotou, Κ.; Ioannou, Ε.; Nomikou, Μ.; Pitterl, F.; Vonaparti, A.; Siapi, E.; Zervou, M.; Roussis, V. An integrated approach using UHPLC-PDA-HRMS and 2D HSQC NMR for the metabolic profiling of the red alga Laurencia: Dereplication and tracing of natural products. Phytochemistry 2014, 108, 208–219. [Google Scholar] [CrossRef]
- Liu, L.; Lu, Y.; Martinez, J.; Bi, Y.; Lian, G.; Wang, T.; Milasta, S.; Wang, J.; Yang, M.; Liu, G.; et al. Proinflammatory signal suppresses proliferation and shifts macrophage metabolism from Myc-dependent to HIF1α-dependent. Proc. Natl. Acad. Sci. USA 2016, 113, 1564–1569. [Google Scholar] [CrossRef]
- Odegaard, J.I.; Chawla, A. The immune system as a sensor of the metabolic state. Immunity 2013, 38, 644–654. [Google Scholar] [CrossRef]
- Vadiveloo, P.K.; Keramidaris, E.; Morrison, W.A.; Stewart, A.G. Lipopolysaccharide-induced cell cycle arrest in macrophages occurs independently of nitric oxide synthase II induction. Biochim. Biophys. Acta 2001, 1539, 140–146. [Google Scholar] [CrossRef] [Green Version]
- Huang, G.-J.; Pan, C.-H.; Wu, C.-H. Sclareol Exhibits Anti-inflammatory Activity in Both Lipopolysaccharide-Stimulated Macrophages and the λ-Carrageenan-Induced Paw Edema Model. J. Nat. Prod. 2012, 75, 54–59. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, Y.-H.; Deng, J.-S.; Pan, H.-P.; Liao, J.-C.; Huang, S.-S.; Huang, G.-J. Sclareol ameliorate lipopolysaccharide-induced acute lung injury through inhibition of MAPK and induction of HO-1 signaling. Int. Immunopharmacol. 2017, 44, 16–25. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, A.; Sica, A.; Sozzani, S.; Allavena, P.; Vecchi, A.; Locati, M. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. 2004, 25, 677–686. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, K.; Hernandez, L.D.; Galá, J.E.; Janeway, C.A.; Medzhitov, R.; Flavell, R.A. IRAK-M Is a Negative Regulator of Toll-like Receptor Signaling. Cell 2002, 110, 191–202. [Google Scholar] [CrossRef] [Green Version]
- Ruffell, D.; Mourkioti, F.; Gambardella, A.; Kirstetter, P.; Lopez, R.G.; Rosenthal, N.; Nerlov, C. A CREB-C/EBPβ cascade induces M2 macrophage- specific gene expression and promotes muscle injury repair. Proc. Natl. Acad. Sci. USA 2009, 106, 17475–17480. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Xu, M.M.; Wang, K.; Adler, A.J.; Vella, A.T.; Zhou Farmington, B.; Zhou, B. Macrophage polarization and meta-inflammation. Transl. Res. 2018, 191, 29–44. [Google Scholar] [CrossRef] [PubMed]
- Lyroni, K.; Patsalos, A.; Daskalaki, M.G.; Doxaki, C.; Soennichsen, B.; Helms, M.; Liapis, I.; Zacharioudaki, V.; Kampranis, S.C.; Tsatsanis, C. Epigenetic and Transcriptional Regulation of IRAK-M Expression in Macrophages. J. Immunol. 2017, 198, 1297–1307. [Google Scholar] [CrossRef]
- Doxaki, C.; Kampranis, S.C.; Eliopoulos, A.G.; Spilianakis, C.; Tsatsanis, C. Coordinated Regulation of miR-155 and miR-146a Genes during Induction of Endotoxin Tolerance in Macrophages. J. Immunol. 2015, 195, 5750–5761. [Google Scholar] [CrossRef] [Green Version]
- Vergadi, E.; Vaporidi, K.; Theodorakis, E.E.; Doxaki, C.; Lagoudaki, E.; Ieronymaki, E.; Alexaki, V.I.; Helms, M.; Kondili, E.; Soennichsen, B.; et al. Akt2 Deficiency Protects from Acute Lung Injury via Alternative Macrophage Activation and miR-146a Induction in Mice. J. Immunol. 2014, 192, 394–406. [Google Scholar] [CrossRef]
- Bradley, M.N.; Zhou, L.; Smale, S.T. C/EBPbeta Regulation in Lipopolysaccharide-Stimulated Macrophages. Mol. Cell. Biol. 2003, 23, 4841–4858. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Liu, X.-j.; Xie, J.; Ma, T.-t.; Meng, X.-m.; Li, J. MiR-146a modulates macrophage polarization by inhibiting Notch1 pathway in RAW264.7 macrophages. Int. Immunopharmacol. 2016, 32, 46–54. [Google Scholar] [CrossRef] [PubMed]
- Vergadi, E.; Vaporidi, K.; Tsatsanis, C. Regulation of Endotoxin Tolerance and Compensatory Anti-inflammatory Response Syndrome by Non-coding RNAs. Front. Immunol. 2018, 9, 2705. [Google Scholar] [CrossRef] [PubMed]
- Androulidaki, A.; Iliopoulos, D.; Arranz, A.; Doxaki, C.; Schworer, S.; Zacharioudaki, V.; Margioris, A.N.; Tsichlis, P.N.; Tsatsanis, C. The Kinase Akt1 Controls Macrophage Response to Lipopolysaccharide by Regulating MicroRNAs. Immunity 2009, 31, 220–231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okayasu, I.; Hatakeyama, S.; Fau-Yamada, M.; Yamada, M.; Fau-Ohkusa, T.; Ohkusa, T.; Fau-Inagaki, Y.; Inagaki, Y.; Fau-Nakaya, R.; Nakaya, R. A novel method in the induction of reliable experimental acute and chronic ulcerative colitis in mice. Gastroenterology 1990, 98, 694–702. [Google Scholar] [CrossRef]
- Jurjus, A.R.; Khoury, N.N.; Reimund, J.-M. Animal models of inflammatory bowel disease. J. Pharmacol. Toxicol. Methods 2004, 50, 81–92. [Google Scholar] [CrossRef] [PubMed]
- Randhawa, P.K.; Singh, K.; Singh, N.; Jaggi, A.S. A review on chemical-induced inflammatory bowel disease models in rodents. Korean J. Physiol. Pharmacol. 2014, 18, 279–288. [Google Scholar] [CrossRef] [PubMed]
- Diaz-Granados, N.; Howe, K.; Lu, J.; McKay, D.M. Dextran sulfate sodium-induced colonic histopathology, but not altered epithelial ion transport, is reduced by inhibition of phosphodiesterase activity. Am. J. Pathol. 2000, 156, 2169–2177. [Google Scholar] [CrossRef]








© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Daskalaki, M.G.; Vyrla, D.; Harizani, M.; Doxaki, C.; Eliopoulos, A.G.; Roussis, V.; Ioannou, E.; Tsatsanis, C.; Kampranis, S.C. Neorogioltriol and Related Diterpenes from the Red Alga Laurencia Inhibit Inflammatory Bowel Disease in Mice by Suppressing M1 and Promoting M2-Like Macrophage Responses. Mar. Drugs 2019, 17, 97. https://0-doi-org.brum.beds.ac.uk/10.3390/md17020097
Daskalaki MG, Vyrla D, Harizani M, Doxaki C, Eliopoulos AG, Roussis V, Ioannou E, Tsatsanis C, Kampranis SC. Neorogioltriol and Related Diterpenes from the Red Alga Laurencia Inhibit Inflammatory Bowel Disease in Mice by Suppressing M1 and Promoting M2-Like Macrophage Responses. Marine Drugs. 2019; 17(2):97. https://0-doi-org.brum.beds.ac.uk/10.3390/md17020097
Chicago/Turabian StyleDaskalaki, Maria G., Dimitra Vyrla, Maria Harizani, Christina Doxaki, Aristides G. Eliopoulos, Vassilios Roussis, Efstathia Ioannou, Christos Tsatsanis, and Sotirios C. Kampranis. 2019. "Neorogioltriol and Related Diterpenes from the Red Alga Laurencia Inhibit Inflammatory Bowel Disease in Mice by Suppressing M1 and Promoting M2-Like Macrophage Responses" Marine Drugs 17, no. 2: 97. https://0-doi-org.brum.beds.ac.uk/10.3390/md17020097