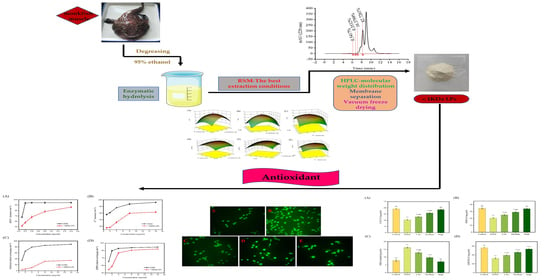
Optimization of Extraction of Bioactive Peptides from Monkfish (Lophius litulon) and Characterization of Their Role in H2O2-Induced Lesion
Abstract
:1. Introduction
2. Results and Discussion
2.1. Selection of the Optimal Enzyme
2.2. Single-Factor Experiments
2.3. Determination of Optimal Conditions Using RSM to Produce LPs by the Action of Neutrase
1.840B2 − 3.42C2
1.420A2 − 2.990B2 − 3.56C2
2.4. MW Distribution of Monkfish Muscle Protein Hydrolysate
2.5. Interception of LPs with Different MW Using Ultrafiltration
2.6. Amino Acid Content of LPs <1 kDa
2.7. Antioxidant Activity of <1 kDa LPs
2.8. Effect of <1 kDa LPs on the Viability of H2O2-Stimulated Cells
2.9. Effects of <1 kDa LPs on the ROS Levels
2.10. Effect of <1 kDa LPs on Antioxidant Activities
3. Materials and Methods
3.1. Pretreatment of Monkfish (Lophius litulon) Muscle
3.2. Optimization of Preparative Conditions of LPs
3.3. Determining the MW Distribution of LPs
3.4. DH and Antioxidant Activity
3.5. Preparation of LPs with Different Molecular Weight
3.6. Effects of <1 kDa LPs on theH2O2-Injured Cells
3.7. Determination of ROS Levels after H2O2 Treatment
3.8. Antioxidant Enzyme Activity of <1 kDa LPs in H2O2-Induced Stress
3.9. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Elias, R.J.; Kellerby, S.S.; Decker, E.A. Antioxidant activity of proteins and peptides. Crit. Rev. Food Sci. Nutr. 2008, 48, 430–441. [Google Scholar] [CrossRef] [PubMed]
- Bernardini, R.D.; Harnedy, P.; Bolton, D.; Kerry, J.; O’Neill, E.; Mullen, A.M.; Hayes, M. Antioxidant and antimicrobial peptidic hydrolysates from muscle protein sources and by-products. Food Chem. 2011, 124, 1296–1307. [Google Scholar] [CrossRef]
- Randy, C.; Tzi, N.; Jack, W. Marine peptides: Bioactivities and applications. Mar. Drugs 2015, 13, 4006–4043. [Google Scholar]
- Bougatef, A.; Hajji, M.; Balti, R.; Lassoued, I.; Triki-Ellouz, Y.; Nasri, M. Antioxidant and free radical-scavenging activities of smooth hound (Mustelus mustelus) muscle protein hydrolysates obtained by gastrointestinal proteases. Food Chem. 2009, 114, 1198–1205. [Google Scholar] [CrossRef]
- Cheung, I.W.Y.; Cheung, L.K.Y.; Tan, N.Y.; Li-Chan, E.C.Y. The role of molecular size in antioxidant activity of peptide fractions from pacific hake (Merluccius productus) hydrolysates. Food Chem. 2012, 134, 1297–1306. [Google Scholar] [CrossRef] [PubMed]
- Sarmadi, B.H.; Ismail, A. Antioxidative peptides from food proteins: A review. Peptides 2010, 31, 1949–1956. [Google Scholar] [CrossRef] [PubMed]
- Rushikesh, S.; Pravin, P.; Seetharama, J. Peptides, peptidomimetics, and polypeptides from marine sources: A wealth of natural sources for pharmaceutical applications. Mar. Drugs 2017, 15, 124–160. [Google Scholar]
- Jonsdottir, R.; Geirsdottir, M.; Hamaguchi, P.Y.; Jamnik, P.; Kristinsson, H.G.; Undeland, I. The ability of in vitro antioxidant assays to predict the efficiency of a cod protein hydrolysate and brown seaweed extract to prevent oxidation in marine food model systems. J. Sci. Food Agric. 2016, 96, 2125–2135. [Google Scholar] [CrossRef]
- Chi, C.; Hu, F.; Wang, B.; Li, Z.; Luo, H. Influence of amino acid compositions and peptide profiles on antioxidant capacities of two protein hydrolysates from skipjack tuna (Katsuwonus pelamis) dark muscle. Mar. Drugs 2015, 13, 2580–2601. [Google Scholar] [CrossRef]
- Borawska, J.; Darewicz, M.; Pliszka, M.; Vegarud, G.E. Antioxidant properties of salmon (Salmo salar L.) protein fraction hydrolysates revealed following their ex vivo digestion and in vitro hydrolysis. J. Sci. Food Agric. 2016, 96, 2764–2772. [Google Scholar] [CrossRef]
- Nazeer, R.A.; Kumar, N.S.S.; Ganesh, R.J. In vitro and in vivo studies on the antioxidant activity of fish peptide isolated from the croaker (Otolithes ruber) muscle protein hydrolysate. Peptides 2012, 35, 261–268. [Google Scholar] [CrossRef]
- Ranathunga, S.; Rajapakse, N.; Kim, S.K. Purification and characterization of antioxidative peptide derived from muscle of conger eel (Conger myriaster). Eur. Food Res. Technol. 2006, 222, 310–315. [Google Scholar] [CrossRef]
- Jaswir, I.; Noviendri, D.; Taher, M.; Mohamed, F.; Octavianti, F.; Lestari, W.; Mukti, A.; Nirwandar, S.; Hamad Almansori, B. Optimization and formulation of fucoxanthin-loaded microsphere (F-LM) using response surface methodology (RSM) and analysis of its fucoxanthin release profile. Molecules 2019, 24, 947. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ovissipour, M.; Kenari, A.A.; Motamedzadegan, A.; Nazari, R.M. Optimization of enzymatic hydrolysis of visceral waste proteins of yellowfin tuna (Thunnus albacares). Food Bioprocess Technol. 2012, 5, 696–705. [Google Scholar] [CrossRef]
- Muhammad, A.S.; Mohammad, Z.; Azizah, A.H.; Nazamid, S. Response surface optimisation for the production of antioxidant hydrolysates from stone fish protein using bromelain. Evid. Based Complem. Altern. Med. 2017, 2017, 1–10. [Google Scholar]
- Yang, S.Y.; Kim, S.W.; Kim, Y.; Lee, S.H.; Jeon, H.; Lee, K.W. Optimization of Maillard reaction with ribose for enhancing anti-allergy effect of fish protein hydrolysates using response surface methodology. Food Chem. 2015, 176, 420–425. [Google Scholar] [CrossRef]
- Gómez, L.J.; Gómez, N.A.; Zapata, J.E.; López-García, G.; Cilla, A.; Alegría, A. Optimization of the red tilapia (Oreochromis spp.) viscera hydrolysis for obtaining iron-binding peptides and evaluation of in vitroiron bioavailability. Foods 2020, 9, 883. [Google Scholar] [CrossRef]
- Chi, C.F.; Wang, B.; Deng, Y.Y.; Wang, Y.M.; Deng, S.G.; Ma, J.Y. Isolation and characterization of three antioxidant pentapeptides from protein hydrolysate of monkfish (Lophius litulon) muscle. Food Res. Int. 2014, 55, 222–228. [Google Scholar] [CrossRef]
- Hu, X.M.; Wang, Y.M.; Zhao, Y.Q.; Chi, C.F.; Wang, B. Antioxidant peptides from the protein hydrolysate of monkfish (Lophius litulon) muscle: Purification, identification, and cytoprotective function on hepg2 cells damage by H2O2. Mar. Drugs 2020, 18, 153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Z.Y.; Zhang, Q.; Li, Y.F.; Dong, L.L.; Liu, S.L. Optimization of ultrasound extraction of Alisma orientalis polysaccharides by response surface methodology and their antioxidant activities. Carbohydr. Polym. 2015, 119, 101–109. [Google Scholar] [CrossRef]
- Raghavan, S.; Kristinsson, H.; Leeuwenburgh, B. Radical scavenging and reducing ability of tilapia (Oreochromis niloticus) protein hydrolysates. J. Agric. Food Chem. 2008, 56, 10359–10367. [Google Scholar] [CrossRef]
- Sonklin, C.; Laohakunjit, N.; Kerdchoechuen, O. Assessment of antioxidant properties of membrane ultrafiltration peptides from mungbean meal protein hydrolysates. PeerJ 2018, 6, 17–36. [Google Scholar] [CrossRef] [PubMed]
- Saidi, S.; Deratani, A.; Belleville, M.; Amar, R.B. Antioxidant properties of peptide fractions from tuna dark muscle protein by-product hydrolysate produced by membrane fractionation process. Food Res. Int. 2014, 65, 329–336. [Google Scholar] [CrossRef]
- Hsu, K. Purification of antioxidative peptides prepared from enzymatic hydrolysates of tuna dark muscle by-product. Food Chem. 2010, 122, 42–48. [Google Scholar] [CrossRef]
- Ahn, C.; Kim, J.; Je, J. Purification and antioxidant properties of octapeptide from salmon byproduct protein hydrolysate by gastrointestinal digestion. Food Chem. 2014, 147, 78–83. [Google Scholar] [CrossRef] [PubMed]
- He, R.; Girgih, A.T.; Malomo, S.A.; Ju, X.; Aluko, R.E. Antioxidant activities of enzymatic rapeseed protein hydrolysates and the membrane ultrafiltration fractions. J. Funct. Foods 2013, 5, 219–227. [Google Scholar] [CrossRef]
- Zheng, J.; Tian, X.; Xu, B.; Yuan, F.; Gong, J.; Yang, Z. Collagen Peptides from Swim Bladders of Giant Croaker (Nibea japonica) and Their Protective Effects against H2O2-Induced Oxidative Damage toward Human Umbilical Vein Endothelial Cells. Mar. Drugs 2020, 18, 430. [Google Scholar] [CrossRef]
- Samaranayaka, A.G.P.; Lichan, E.C.Y. Food-derived peptidic antioxidants: A review of their production, assessment, and potential applications. J. Funct. Foods 2011, 3, 229–254. [Google Scholar] [CrossRef]
- Udenigwe, C.C.; Aluko, R.E. Chemometric analysis of the amino acid requirements of antioxidant food protein hydrolysates. Int. J. Mol. Sci. 2011, 12, 3148–3161. [Google Scholar] [CrossRef] [Green Version]
- Xiao, X.; Liu, J.; Hu, J.; Zhu, X.; Yang, H.; Wang, C.; Zhang, Y. Protective effects of protopine on hydrogen peroxide-induced oxidative injury of pc12 cells via Ca2+ antagonism and antioxidant mechanisms. Eur. J. Pharmacol. 2008, 591, 21–27. [Google Scholar] [CrossRef]
- Hu, X.; Li, W.; Kong, X.; Han, H.; Tang, Y.; Yu, F.; Yang, Z.; Ding, G. Optimization of extraction technology of immunologically active peptides from Nibea japonica by response surface methodology. Sci. Technol. Food Ind. 2019, 17, 173–178. [Google Scholar]
- Hu, Z.; Yang, P.; Zhou, C.; Li, S.; Hong, P. Marine collagen peptides from the skin of Nile tilapia (Oreochromis niloticus): Characterization and wound healing evaluation. Mar. Drugs 2017, 15, 102. [Google Scholar] [CrossRef]
- Wu, Z.; Ding, G.; Huang, F.; Yang, Z.; Yu, F.; Tang, Y.; Jia, Y.; Zheng, Y.; Chen, R. Anticancer activity of Anthopleura anjunae oligopeptides in prostate cancer du-145 cells. Mar. Drugs 2018, 16, 125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Noman, A.; Xu, Y.; Al-Bukhaiti, W.Q.; Abed, S.M.; Ali, A.H.; Ramadhan, A.H.; Xia, W. Influence of enzymatic hydrolysis conditions on the degree of hydrolysis and functional properties of protein hydrolysate obtained from Chinese sturgeon (Acipenser sinensis) by using papain enzyme. Process Biochem. 2018, 67, 19–28. [Google Scholar] [CrossRef]
- Horwitz, W. Official methods of analysis of AOAC international. Trends Food Sci. Technol. 1995, 6, 382. [Google Scholar]
- Chen, Y.; Jin, H.; Yang, F.; Jin, S.; Liu, C.; Zhang, L.; Huang, J.; Wang, S.; Yan, Z.; Cai, X. Physicochemical, antioxidant properties of giant croaker (Nibea japonica) swim bladders collagen and wound healing evaluation. Int. J. Biol. Macromol. 2019, 138, 483–491. [Google Scholar] [CrossRef]
- Tang, Y.; Jin, S.; Li, X.; Li, X.; Hu, X.; Chen, Y.; Huang, F.; Yang, Z.; Yu, F.; Ding, G. Physicochemical properties and biocompatibility evaluation of collagen from the skin of giant croaker (Nibea japonica). Mar. Drugs 2018, 16, 222. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Li, J.; Lin, S.; Yang, Z.; Jin, H. Preparation of antioxidant peptide by microwave-assisted hydrolysis of collagen and its protective effect against H2O2-induced damage of raw264.7 cells. Mar. Drugs 2019, 17, 642. [Google Scholar] [CrossRef] [Green Version]
- Takamiya, R.; Fukunaga, K.; Arita, M.; Miyata, J.; Seki, H.; Minematsu, N.; Suematsu, M.; Asano, K. Resolvin e1 maintains macrophage function under cigarette smoke-induced oxidative stress. FEBS Open Bio 2012, 2, 328–333. [Google Scholar] [CrossRef] [Green Version]









| Variables | Sum of Squares | df | Mean Square | F-Value | p-Value |
|---|---|---|---|---|---|
| A (material/water ratio) | 2.92 | 1 | 2.92 | 8.99 | 0.0200 |
| B (extraction time) | 1.73 | 1 | 1.73 | 5.32 | 0.0545 |
| C (PH) | 0.017 | 1 | 0.017 | 0.053 | 0.8242 |
| AB | 0.31 | 1 | 0.31 | 0.96 | 0.3604 |
| AC | 0.55 | 1 | 0.55 | 1.70 | 0.2333 |
| BC | 0.035 | 1 | 0.035 | 0.11 | 0.7538 |
| A2 | 60.52 | 1 | 60.52 | 186.41 | <0.0001 |
| B2 | 14.25 | 1 | 14.25 | 43.90 | 0.0003 |
| C2 | 49.23 | 1 | 49.23 | 151.65 | <0.0001 |
| Model | 142.64 | 9 | 15.85 | 48.82 | <0.0001 |
| Lack of fit | 4.02 | 7 | 0.57 | ||
| Residual | 1.55 | 3 | 0.52 | 2.88 | 0.1663 |
| Pure error | 0.72 | 4 | 0.18 | ||
| Cor. total | 144.91 | 16 | |||
| R2 = 0.9843 | RAdj2 = 0.9642 | CV = 3.49% | |||
| Variables | Sum of Squares | df | Mean Square | F-Value | p-Value |
|---|---|---|---|---|---|
| A (material/water ratio) | 5.43 | 1 | 5.43 | 6.40 | 0.0291 |
| B (extraction time) | 6.35 | 1 | 6.35 | 7.48 | 0.0393 |
| C (PH) | 0.48 | 1 | 0.48 | 0.57 | 0.4757 |
| AB | 0.84 | 1 | 0.84 | 0.99 | 0.3539 |
| AC | 5.78 | 1 | 5.78 | 6.80 | 0.2701 |
| BC | 1.22 | 1 | 1.22 | 1.43 | 0.0350 |
| A^2 | 37.56 | 1 | 37.56 | 44.24 | 0.0161 |
| B^2 | 8.43 | 1 | 8.43 | 9.93 | 0.0003 |
| C^2 | 53.45 | 1 | 53.45 | 62.95 | <0.0001 |
| Model | 129.48 | 9 | 14.39 | 16.95 | 0.0006 |
| Lack of fit | 5.94 | 7 | 0.85 | ||
| Residual | 4.87 | 3 | 1.62 | 6.06 | 0.0571 |
| Pure error | 1.07 | 4 | 0.27 | ||
| Cor. total | 135.42 | 16 | |||
| R2 = 0.9561 | RAdj2 = 0.8997 | CV = 1.04% | |||
| Amino Acids | LPs |
|---|---|
| Asp | 5.82 ± 0.190 |
| Thr | 2.83 ± 0.083 |
| Ser | 2.89 ± 0.080 |
| Glu | 9.75 ± 0.297 |
| Gly | 2.53 ± 0.080 |
| Ala | 3.45 ± 0.107 |
| Cys | 0.00 |
| Val | 3.51 ± 0.096 |
| Met | 2.42 ± 0.051 |
| Lle | 2.79 ± 0.079 |
| Leu | 5.57 ± 0.152 |
| Tyr | 1.81 ± 0.014 |
| Phe | 6.19 ± 0.079 |
| Lys | 5.48 ± 0.167 |
| His | 1.55 ± 0.102 |
| Arg | 3.70 ± 0.105 |
| Pro | 0.00 |
| HAA | 25.74 |
| PCAA | 10.73 |
| NCAA | 15.59 |
| EAA | 28.43 |
| Types of Protease | Optimum Temperature (°C) | Optimum pH |
|---|---|---|
| neutrase | 45 °C | 7.0 |
| alcalase | 45 °C | 10.0 |
| papain | 60 °C | 6.0 |
| pepsin | 37 °C | 2.0 |
| trypsin | 37 °C | 8.0 |
| Variables | Levels and Range | ||
|---|---|---|---|
| −1 | 0 | 1 | |
| A muscle/water ratio (g·mL−1) | 1:4 | 1:5 | 1:6 |
| B extraction time (h) | 4 | 5 | 6 |
| C pH | 6.5 | 7 | 7.5 |
| Run Numbers | A Extraction Time (h) | B Muscle/Water Ratio (g/mL) | C PH | DH% | DPPH/% |
|---|---|---|---|---|---|
| 1 | −1 | 0 | −1 | 13.0088 | 86.7955 |
| 2 | 0 | 0 | 0 | 20.4425 | 93.4659 |
| 3 | 0 | −1 | −1 | 15.2389 | 86.9091 |
| 4 | −1 | 0 | 1 | 11.8939 | 86.375 |
| 5 | 0 | 0 | 0 | 20.0708 | 92.1818 |
| 6 | 1 | −1 | 0 | 15.6106 | 88.1136 |
| 7 | 0 | 0 | 0 | 20.8142 | 92.4659 |
| 8 | 0 | 1 | −1 | 14.8673 | 87.0568 |
| 9 | 0 | 0 | 0 | 21.1858 | 92.4091 |
| 10 | −1 | −1 | 0 | 15.6106 | 87.7386 |
| 11 | 1 | 0 | 1 | 14.4956 | 89.752 |
| 12 | 1 | 1 | 0 | 14.8673 | 89.7727 |
| 13 | −1 | 1 | 0 | 13.7522 | 87.5682 |
| 14 | 1 | 0 | −1 | 14.1239 | 87.9659 |
| 15 | 0 | 1 | 1 | 15.2389 | 87.7955 |
| 16 | 0 | −1 | 1 | 15.9823 | 82.841 |
| 17 | 0 | 0 | 0 | 20.4425 | 92.9773 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tian, X.; Zheng, J.; Xu, B.; Ye, J.; Yang, Z.; Yuan, F. Optimization of Extraction of Bioactive Peptides from Monkfish (Lophius litulon) and Characterization of Their Role in H2O2-Induced Lesion. Mar. Drugs 2020, 18, 468. https://0-doi-org.brum.beds.ac.uk/10.3390/md18090468
Tian X, Zheng J, Xu B, Ye J, Yang Z, Yuan F. Optimization of Extraction of Bioactive Peptides from Monkfish (Lophius litulon) and Characterization of Their Role in H2O2-Induced Lesion. Marine Drugs. 2020; 18(9):468. https://0-doi-org.brum.beds.ac.uk/10.3390/md18090468
Chicago/Turabian StyleTian, Xiaoxiao, Jiawen Zheng, Baogui Xu, Jiena Ye, Zuisu Yang, and Falei Yuan. 2020. "Optimization of Extraction of Bioactive Peptides from Monkfish (Lophius litulon) and Characterization of Their Role in H2O2-Induced Lesion" Marine Drugs 18, no. 9: 468. https://0-doi-org.brum.beds.ac.uk/10.3390/md18090468