Biosynthesis of Polyketides in Streptomyces
Abstract
:1. Introduction
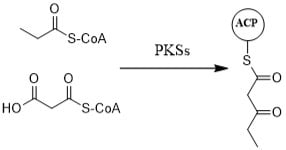
2. Polyketide Synthases Type I
2.1. Biosynthesis of Rapamycin
2.2. Biosynthesis of Avermectin
2.3. Biosynthesis of Candicidin
2.4. Biosynthesis of Tautomycetin
3. Polyketide Synthases Type II
3.1. Biosynthesis of Doxorubicin
3.2. Biosynthesis of Medermycin
3.3. Biosynthesis of Hedamycin
3.4. Biosynthesis of Fredericamycin
4. Polyketide Synthases Type III
4.1. Biosynthesis of Germicidin
4.2. Biosynthesis of Tetrahydroxynaphthalene
4.3. Biosynthesis of Dihydroxyphenylglycine
4.4. Biosynthesis of Alkylresorcinol
5. Conclusions
Acknowledgments
Conflicts of Interest
References
- Moore, B.S.; Hopke, J.N. Discovery of a new bacterial polyketide biosynthetic pathway. ChemBioChem 2001, 2, 35–38. [Google Scholar] [CrossRef]
- Rokem, J.S.; Lantz, A.E.; Nielsen, J. Systems biology of antibiotic production by microorganisms. Nat. Prod. Rep. 2007, 24, 1262. [Google Scholar] [CrossRef]
- Katsuyama, Y.; Funa, N.; Miyahisa, I.; Horinouchi, S. Synthesis of unnatural flavonoids and stilbenes by exploiting the plant biosynthetic pathway in Escherichia coli. Chem. Biol. 2007, 14, 613–621. [Google Scholar] [CrossRef]
- Chopra, I.; Roberts, M. Tetracycline antibiotics: Mode of action, applications, molecular biology, and epidemiology of bacterial resistance tetracycline antibiotics: Mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 2001, 65, 232–260. [Google Scholar] [CrossRef] [PubMed]
- Ghannoum, M.A.; Rice, L.B. Antifungal agents: Mode of action, mechanisms of resistance, and correlation of these mechanisms with bacterial resistance. Clin. Microbiol. Rev. 1999, 12, 501–517. [Google Scholar] [CrossRef]
- Tacar, O.; Sriamornsak, P.; Dass, C.R. Doxorubicin: An update on anticancer molecular action, toxicity and novel drug delivery systems. J. Pharm. Pharmacol. 2013, 65, 157–170. [Google Scholar] [CrossRef]
- Shushni, M.A.M.; Singh, R.; Mentel, R.; Lindequist, U. Balticolid: A new 12-membered macrolide with antiviral activity from an Ascomycetous fungus of marine origin. Mar. Drugs 2011, 9, 844–851. [Google Scholar] [CrossRef]
- Li, J.; Kim, S.G.; Blenis, J. Rapamycin: One drug, many effects. Cell Metab. 2014, 19, 373–379. [Google Scholar] [CrossRef] [PubMed]
- Van de Donk, N.W.C.J.; Kamphuis, M.M.J.; Lokhorst, H.M.; Bloem, A.C. The cholesterol lowering drug lovastatin induces cell death in myeloma plasma cells. Leukemia 2002, 16, 1362–1371. [Google Scholar] [CrossRef]
- Hleba, L.; Charousova, I.; Cisarova, M.; Kovacik, A.; Kormanec, J.; Medo, J.; Bozik, M.; Javorekova, S. Rapid identification of Streptomyces tetracycline producers by MALDI-TOF mass spectrometry. J. Environ. Sci. Heal. Part A 2018. [Google Scholar] [CrossRef] [PubMed]
- Parthasarathy, R.; Sathiyabama, M. Lovastatin-producing endophytic fungus isolated from a medicinal plant Solanum xanthocarpum. Nat. Prod. Res. 2015, 29, 2282–2286. [Google Scholar] [CrossRef]
- Huang, Q.; Lu, G.; Shen, H.-M.; Chung, M.C.M.; Ong, C.N. Anti-cancer properties of anthraquinones from rhubarb. Med. Res. Rev. 2006, 27, 609–630. [Google Scholar] [CrossRef]
- Kohli, G.S.; John, U.; Figueroa, R.I.; Rhodes, L.L.; Harwood, D.T.; Groth, M.; Bolch, C.J.S.; Murray, S.A. Polyketide synthesis genes associated with toxin production in two species of Gambierdiscus (Dinophyceae). BMC Genomics 2015, 16, 410. [Google Scholar] [CrossRef]
- Florian, P.; Monika, H. Polyketides in insects: Ecological role of these widespread chemicals and evolutionary aspects of their biogenesis. Biol. Rev. 2008, 83, 209–226. [Google Scholar]
- Adele, C.; Guido, C.; Guido, V.; Angelo, F. Shaping the polypropionate biosynthesis in the solar-powered mollusc Elysia viridis. ChemBioChem 2008, 10, 315–322. [Google Scholar]
- Müller, R.; Wink, J. Future potential for anti-infectives from bacteria—How to exploit biodiversity and genomic potential. Int. J. Med. Microbiol. 2014, 304, 3–13. [Google Scholar] [CrossRef]
- Widyastuti, Y.; Lisdiyanti, P.; Ratnakomala, S.; Kartina, G.; Ridwan, R.; Rohmatussolihat, R.; Rosalinda Prayitno, N.; Triana, E.; Widhyastuti, N.; et al. Genus diversity of Actinomycetes in Cibinong Science Center, West Java, Indonesia. Microbiol. Indones. 2012, 6, 165–172. [Google Scholar] [CrossRef]
- Lucas, X.; Senger, C.; Erxleben, A.; Grüning, B.A.; Döring, K.; Mosch, J.; Flemming, S.; Günther, S. StreptomeDB: A resource for natural compounds isolated from Streptomyces species. Nucleic Acids Res. 2013, 41, D1130–D1136. [Google Scholar] [CrossRef]
- Igarashi, M.; Takahashi, Y.; Shitara, T.; Nakamura, H.; Naganawa, H.; Miyake, T.; Akamatsu, Y. Caprazamycins, novel lipo-nucleoside antibiotics, from Streptomyces sp. J. Antibiot. 2005, 58, 327–337. [Google Scholar] [CrossRef]
- Dutta, S.; Basak, B.; Bhunia, B.; Chakraborty, S.; Dey, A. Kinetics of rapamycin production by Streptomyces hygroscopicus MTCC 4003. 3 Biotech. 2014, 4, 523–531. [Google Scholar] [CrossRef]
- Rodríguez, L.; Rodríguez, D.; Olano, C.; Braña, A.F.; Méndez, C.; Salas, J.A. Functional analysis of OleY L-oleandrosyl 3-O-methyltransferase of the oleandomycin biosynthetic pathway in Streptomyces antibioticus. J. Bacteriol. 2001, 183, 5358–5363. [Google Scholar] [CrossRef] [PubMed]
- Elibol, M. Optimization of medium composition for actinorhodin production by Streptomyces coelicolor A3(2) with response surface methodology. Process Biochem. 2004, 39, 1057–1062. [Google Scholar] [CrossRef]
- Pokhrel, A.R.; Chaudhary, A.K.; Nguyen, H.T.; Dhakal, D.; Le, T.T.; Shrestha, A.; Liou, K.; Sohng, J.K. Overexpression of a pathway specific negative regulator enhances production of daunorubicin in bldA deficient Streptomyces peucetius ATCC 27952. Microbiol. Res. 2016, 192, 96–102. [Google Scholar] [CrossRef]
- Shelest, E.; Heimerl, N.; Fichtner, M.; Sasso, S. Multimodular type I polyketide synthases in algae evolve by module duplications and displacement of AT domains in trans. BMC Genomics 2015, 16, 1–15. [Google Scholar] [CrossRef]
- Hopwood, D.A. Cracking the polyketide code. PLoS Biol. 2004, 2, 166–169. [Google Scholar] [CrossRef]
- Staunton, J.; Weissman, K.J. Polyketide biosynthesis: A millennium review. Nat. Prod. Rep. 2001, 18, 380–416. [Google Scholar] [CrossRef]
- Lal, R.; Kumari, R.; Kaur, H.; Khanna, R.; Dhingra, N.; Tuteja, D. Regulation and manipulation of the gene clusters encoding type I PKSs. Trends Biotechnol. 2000, 18, 264–274. [Google Scholar] [CrossRef]
- Okamoto, S.; Taguchi, T.; Ochi, K.; Ichinose, K. Biosynthesis of actinorhodin and related antibiotics: Discovery of alternative routes for quinone formation encoded in the act gene cluster. Chem. Biol. 2009, 16, 226–236. [Google Scholar] [CrossRef] [PubMed]
- Funabashi, M.; Funa, N.; Horinouchi, S. Phenolic lipids synthesized by type III polyketide synthase confer penicillin resistance on Streptomyces griseus. J. Biol. Chem. 2008, 283, 13983–13991. [Google Scholar] [CrossRef]
- Vance, S.; Tkachenko, O.; Thomas, B.; Bassuni, M.; Hong, H.; Nietlispach, D.; Broadhurst, W. Sticky swinging arm dynamics: Studies of an acyl carrier protein domain from the mycolactone polyketide synthase. Biochem. J. 2016, 473, 1097–1110. [Google Scholar] [CrossRef] [PubMed]
- Ruan, X.; Stassi, D.; Lax, S.A.; Katz, L. A second type I PKS gene cluster isolated from Streptomyces hygroscopicus ATCC 29253, a rapamycin-producing strain. Gene 1997, 203, 1–9. [Google Scholar] [CrossRef]
- Choi, S.S.; Hur, Y.A.; Sherman, D.H.; Kim, E.S. Isolation of the biosynthetic gene cluster for tautomycetin, a linear polyketide T cell-specific immunomodulator from Streptomyces sp. CK4412. Microbiology 2007, 153, 1095–1102. [Google Scholar] [CrossRef] [PubMed]
- Mazzei, T.; Mini, E.; Noveffi, A.; Periti, P. Chemistry and mode of action of macrolides. J. Antimicrob. Chemother. 1993, 31, 1–9. [Google Scholar] [CrossRef]
- Schwecke, T.; Aparicio, J.F.; Molnár, I.; König, A.; Khaw, L.E.; Haydock, S.F.; Oliynyk, M.; Caffrey, P.; Cortés, J.; Lester, J.B. The biosynthetic gene cluster for the polyketide immunosuppressant rapamycin. Proc. Natl. Acad. Sci. USA 1995, 92, 7839–7843. [Google Scholar] [CrossRef] [PubMed]
- Motamedi, H.; Shafiee, A. The biosynthetic gene cluster for the macrolactone ring of the immunosuppressant FK506. Eur. J. Biochem. 1998, 256, 528–534. [Google Scholar] [CrossRef]
- Karray, F.; Darbon, E.; Oestreicher, N.; Dominguez, H.; Tuphile, K.; Gagnat, J.; Blondelet-Rouault, M.H.; Gerbaud, C.; Pernodet, J.L. Organization of the biosynthetic gene cluster for the macrolide antibiotic spiramycin in Streptomyces ambofaciens. Microbiology 2007, 153, 4111–4122. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Zhao, L.; Liu, H.W.; Sherman, D.H. A gene cluster for macrolide antibiotic biosynthesis in Streptomyces venezuelae: Architecture of metabolic diversity. Proc. Natl. Acad. Sci. USA 1998, 95, 12111–12116. [Google Scholar] [CrossRef]
- Cruz, M.C.; Cavallo, L.M.; Görlach, J.M.; Cox, G.; Perfect, J.R.; Cardenas, M.E.; Heitman, J. Rapamycin antifungal action is mediated via conserved complexes with FKBP12 and TOR kinase homologs in Cryptococcus neoformans. Mol. Cell Biol. 1999, 19, 4101–4112. [Google Scholar] [CrossRef]
- Bastidas, R.J.; Shertz, C.A.; Lee, S.C.; Heitman, J.; Cardenas, M.E. Rapamycin exerts antifungal activity in vitro and in vivo against Mucor circinelloides via FKBP12-dependent inhibition of tor. Eukaryot. Cell 2012, 11, 270–281. [Google Scholar] [CrossRef]
- Kwan, D.H.; Schulz, F. The stereochemistry of complex polyketide biosynthesis by modular polyketide synthases. Molecules 2011, 16, 6092–6115. [Google Scholar] [CrossRef]
- Ikeda, H.; Nonomiya, T.; Usami, M.; Ohta, T.; Omura, S. Organization of the biosynthetic gene cluster for the polyketide anthelmintic macrolide avermectin in Streptomyces avermitilis. Proc. Natl. Acad. Sci. USA 1999, 96, 9509–9514. [Google Scholar] [CrossRef] [PubMed]
- Ward, S.L.; Hu, Z.; Schirmer, A.; Reid, R.; Revill, W.P.; Reeves, C.D.; Petrakovsky, O.V.; Dong, S.D.; Katz, L. Chalcomycin biosynthesis gene cluster from Streptomyces bikiniensis: Novel features of an unusual ketolide produced through expression of the chm polyketide synthase in Streptomyces fradiae. Antimicrob. Agents Chemother. 2004, 48, 4703–4712. [Google Scholar] [CrossRef]
- Gil, J.A.; Campelo-Diez, A.B. Candicidin biosynthesis in Streptomyces griseus. Appl. Microbiol. Biotechnol. 2003, 60, 633–642. [Google Scholar] [CrossRef]
- Campelo, A.B.; Gil, J.A. The candicidin gene cluster from Streptomyces griseus IMRU 3570. Microbiology 2002, 148, 51–59. [Google Scholar] [CrossRef]
- Kino, T.; Hatanaka, H.; Hashimoto, M.; Nishiyama, M.; Goto, T.; Okuhara, M.; Kohsaka, M.; Aoki, H.; Imanaka, H. FK-506, a novel immunosuppressant isolated from a Streptomyces. I. Fermentation, isolation, and physico-chemical and biological characteristics. J. Antibiot. (Tokyo) 1987, 40, 1249–1255. [Google Scholar] [CrossRef]
- Wu, K.; Chung, L.; Revill, W.P.; Katz, L.; Reeves, C.D. The FK520 gene cluster of Streptomyces hygroscopicus var. ascomyceticus (ATCC 14891) contains genes for biosynthesis of unusual polyketide extender units. Gene 2000, 251, 81–90. [Google Scholar] [CrossRef]
- Aparicio, J.; Colina, A.; Ceballos, E.; Martin, J. The biosynthetic gene cluster for the 26-membered ring polyene macrolide pimaricin. A new polyketide synthase organization encoded by two subclusters separated by functionalization genes. J. Biol. Chem. 1999, 274, 10133–10139. [Google Scholar] [CrossRef]
- Fiers, W.D.; Dodge, G.J.; Li, Y.; Smith, J.L.; Fecik, R.A.; Aldrich, C.C. Tylosin polyketide synthase module 3: Stereospecificity, stereoselectivity and steady-state kinetic analysis of β-processing domains via diffusible, synthetic substrates. Chem. Sci. 2015, 6, 5027–5033. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, Z.; Li, M.; Wen, Y.; Song, Y.; Li, J. Construction of ivermectin producer by domain swaps of avermectin polyketide synthase in Streptomyces avermitilis. Appl. Microbiol. Biotechnol. 2006, 72, 986–994. [Google Scholar] [CrossRef]
- Hammond, S.M.; Kliger, B.N. Mode of action of the polyene antibiotic candicidin: Binding factors in the wall of Candida albicans. Antimicrob. Agents Chemother. 1976, 9, 561–568. [Google Scholar] [CrossRef]
- Cheng, X.-C.; Kihara, T.; Ying, X.; Uramoto, M.; Osada, H.; Kusakabe, H.; Wang, B.-N.; Kobayashi, Y.; Ko, K.; Yamaguchi, I.; Shen, Y.-C.; Isono, K. A new antibiotic, tautomycetin. J. Antibiot. 1989, 42, 141–144. [Google Scholar]
- Hertweck, C.; Luzhetskyy, A.; Rebets, Y.; Bechthold, A. Type II polyketide synthases: Gaining a deeper insight into enzymatic teamwork. Nat. Prod. Rep. 2007, 24, 162–190. [Google Scholar] [CrossRef]
- Malla, S.; Prasad Niraula, N.; Singh, B.; Liou, K.; Kyung Sohng, J. Limitations in doxorubicin production from Streptomyces peucetius. Microbiol. Res. 2010, 165, 427–435. [Google Scholar] [CrossRef]
- Jakeman, D.L.; Bandi, S.; Graham, C.L.; Reid, T.R.; Wentzell, J.R.; Douglas, S.E. Antimicrobial activities of jadomycin B and structurally related analogues. Antimicrob. Agents Chemother. 2009, 53, 1245–1247. [Google Scholar] [CrossRef]
- Lombó, F.; Menéndez, N.; Salas, J.A.; Méndez, C. The aureolic acid family of antitumor compounds: Structure, mode of action, biosynthesis, and novel derivatives. Appl. Microbiol. Biotechnol. 2006, 73, 1–14. [Google Scholar] [CrossRef]
- Petkovic, H.; Cullum, J.; Hranueli, D.; Hunter, I.S.; Peric-Concha, N.; Pigac, J.; Thamchaipenet, A.; Vujaklija, D.; Long, P.F. Genetics of Streptomyces rimosus, the oxytetracycline producer. Microbiol. Mol. Biol. Rev. 2006, 70, 704–728. [Google Scholar] [CrossRef] [PubMed]
- Shen, B.; Hutchinson, C.R. Triple hydroxylation of tetracenomycin A2 to tetracenomycin C in Streptomyces glaucescens. Overexpression of the tcmG gene in Streptomyces lividans and characterization of the tetracenomycin A2 oxygenase. J. Biol. Chem. 1994, 1326, 30726–30733. [Google Scholar]
- Zhang, Z.; Pan, H.-X.; Tang, G.-L. New insights into bacterial type II polyketide biosynthesis. F1000Research 2017, 6, 172. [Google Scholar] [CrossRef] [PubMed]
- Komaki, H.; Harayama, S. Sequence diversity of type II polyketide synthase genes in Streptomyces. Actinomycetologica 2006, 20, 42–48. [Google Scholar] [CrossRef]
- Chan, Y.A.; Podevels, A.M.; Kevany, B.M.; Thomas, M.G. Biosynthesis of polyketide synthase extender units. Nat. Prod. Rep. 2009, 26, 90–114. [Google Scholar] [CrossRef] [PubMed]
- Minotti, G. Anthracyclines: Molecular advances and pharmacologic developments in antitumor activity and cardiotoxicity. Pharmacol. Rev. 2004, 56, 185–229. [Google Scholar] [CrossRef]
- Yang, F.; Teves, S.S.; Kemp, C.J.; Henikoff, S. Doxorubicin, DNA torsion, and chromatin dynamics. Biochim. Biophys. Acta 2014, 1845, 84–89. [Google Scholar] [CrossRef] [PubMed]
- Thorn, C.F.; Oshiro, C.; Marsh, S.; Hernandez-Boussard, T.; McLeod, H.; Klein, T.E.; Altman, R.B. Doxorubicin pathways: Pharmacodynamics and adverse effects. Pharmacogenet. Genomics 2011, 21, 440–446. [Google Scholar] [CrossRef] [PubMed]
- Bao, W.; Sheldon, P.J.; Wendt-Pienkowski, E.; Richard Hutchinson, C. The Streptomyces peucetius dpsC gene determines the choice of starter unit in biosynthesis of the daunorubicin polyketide. J. Bacteriol. 1999, 181, 4690–4695. [Google Scholar]
- Grimm, A.; Madduri, K.; Ali, A.; Hutchinson, C.R. Characterization of the Streptomyces peucetius ATCC 29050 genes encoding doxorubicin polyketide synthase. Gene 1994, 151, 1–10. [Google Scholar] [CrossRef]
- Rajgarhia, V.B.; Strohl, W.R. Minimal Streptomyces sp. strain C5 daunorubicin polyketide biosynthesis genes required for aklanonic acid biosynthesis. J. Bacteriol. 1997, 179, 2690–2696. [Google Scholar] [CrossRef]
- Hutchinson, C.R. Biosynthetic studies of daunorubicin and tetracenomycin C. Chem. Rev. 1997, 97, 2525–2536. [Google Scholar] [CrossRef]
- Lomovskaya, N.; Doi-Katayama, Y.; Filippini, S.; Nastro, C.; Fonstein, L.; Gallo, M.; Colombo, A.L.; Hutchinson, C.R. The Streptomyces peucetius dpsY and dnrX genes govern early and late steps of daunorubicin and doxorubicin biosynthesis. J. Bacteriol. 1998, 180, 2379–2386. [Google Scholar]
- Ichinose, K.; Ozawa, M.; Itou, K.; Kunieda, K.; Ebizuka, Y. Cloning, sequencing and heterologous expression of the medermycin biosynthetic gene cluster of Streptomyces sp. AM-7161: Towards comparative analysis of the benzoisochromanequinone gene clusters. Microbiology 2003, 149, 1633–1645. [Google Scholar] [CrossRef] [PubMed]
- Takano, S.; Hasuda, K.; Ito, A.; Koide, Y.; Ishii, F. A new antibiotic, medermycin. J. Antibiot. (Tokyo) 1976, 29, 765–768. [Google Scholar] [CrossRef]
- Pickens, L.B.; Tang, Y. Oxytetracycline biosynthesis. J. Biol. Chem. 2010, 285, 27509–27515. [Google Scholar] [CrossRef]
- Fischer, C.; Lipata, F.; Rohr, J. The complete gene cluster of the antitumor agent gilvocarcin V and its implication for the biosynthesis of the gilvocarcins. J. Am. Chem. Soc. 2003, 125, 7818–7819. [Google Scholar] [CrossRef] [PubMed]
- Lombó, F.; Abdelfattah, M.S.; Braça, A.F.; Salas, J.A. Elucidation of oxygenation steps during oviedomycin biosynthesis and generation of derivatives with increased antitumor activity. ChemBioChem 2009, 10, 296–303. [Google Scholar] [CrossRef]
- Xu, Z.; Jakobi, K.; Welzel, K.; Hertweck, C. Biosynthesis of the antitumor agent chartreusin involves the oxidative rearrangement of an anthracyclic polyketide. Chem. Biol. 2005, 12, 579–588. [Google Scholar] [CrossRef] [PubMed]
- Bretschneider, T.; Zocher, G.; Unger, M.; Scherlach, K.; Stehle, T.; Hertweck, C. A ketosynthase homolog uses malonyl units to form esters in cervimycin biosynthesis. Nat. Chem. Biol. 2011, 8, 154–161. [Google Scholar] [CrossRef] [PubMed]
- Herold, K.; Xu, Z.; Gollmick, F.A.; Gräfe, U.; Hertweck, C. Biosynthesis of cervimycin C, an aromatic polyketide antibiotic bearing an unusual dimethylmalonyl moiety. Organ. Biomol. Chem. 2004, 2, 2411–2414. [Google Scholar] [CrossRef]
- Jakobi, K.; Hertweck, C. A gene cluster encoding resistomycin biosynthesis in Streptomyces resistomycificus; exploring polyketide cyclization beyond linear and angucyclic patterns. J. Am. Chem. Soc. 2004, 126, 2298–2299. [Google Scholar] [CrossRef] [PubMed]
- Menendez, N.; Nur-e-Alam, M.; Bran, A.F.; Rohr, J.; Salas, J.A.; Mendez, C. Biosynthesis of the antitumor chromomycin A3 in Streptomyces griseus: Analysis of the gene cluster and rational design of novel chromomycin analogs. Chem. Biol. 2004, 11, 21–32. [Google Scholar]
- Sun, L.; Zeng, J.; Cui, P.; Wang, W.; Yu, D.; Zhan, J. Manipulation of two regulatory genes for efficient production of chromomycins in Streptomyces reseiscleroticus. J. Biol. Eng. 2018, 12, 1–11. [Google Scholar] [CrossRef]
- Das, A.; Khosla, C. In vivo and in vitro analysis of the hedamycin polyketide synthase. Chem. Biol. 2009, 16, 1197–1207. [Google Scholar] [CrossRef]
- Bililign, T.; Hyun, C.-G.; Williams, J.S.; Czisny, A.M.; Thorson, J.S. The hedamycin locus implicates a novel aromatic PKS priming mechanism. Chem. Biol. 2004, 11, 959–969. [Google Scholar] [CrossRef]
- Das, A.; Szu, P.; Fitzgerald, J.T.; Khosla, C. Mechanism and engineering of polyketide chain initiation in fredericamycin biosynthesis. J. Am. Chem. Soc. 2010, 132, 8831–8833. [Google Scholar] [CrossRef]
- Chen, Y.; Wendt-Pienkowski, E.; Shen, B. Identification and utility of FdmR1 as a Streptomyces antibiotic regulatory protein activator for fredericamycin production in Streptomyces griseus ATCC 49344 and heterologous hosts. J. Bacteriol. 2008, 190, 5587–5596. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, A.; Fukamachi, N.; Yamaki, K.; Hayashi, M.; Oh-ishi, S.; Kobayashi, B.; Omura, S. Inhibition of platelet aggregation by medermycin and it’s related isochromanequinone antibiotics. J. Antibiot. 1987, 40, 1075–1076. [Google Scholar] [CrossRef]
- Tu, L.C.; Melendy, T.; Beerman, T.A. DNA damage responses triggered by a highly cytotoxic monofunctional DNA alkylator, hedamycin, a pluramycin antitumor antibiotic. Mol. Cancer Ther. 2004, 3, 577–585. [Google Scholar] [PubMed]
- Szu, P.-H.; Govindarajan, S.; Meehan, M.J.; Das, A.; Nguyen, D.D.; Dorrestein, P.C.; Minshull, J.; Khosla, C. Analysis of the ketosynthase-chain length factor heterodimer from the fredericamycin polyketide synthase. Acta Neurobiol. Exp. (Wars) 2011, 18, 1021–1031. [Google Scholar] [CrossRef]
- Shen, B. Polyketide biosynthesis beyond the type I, II and III polyketide synthase paradigms. Curr. Opin. Chem. Biol. 2003, 7, 285–295. [Google Scholar] [CrossRef]
- Yu, D.; Xu, F.; Zeng, J.; Zhan, J. Type III polyketide synthases in natural product biosynthesis. IUBMB Life 2012, 64, 285–295. [Google Scholar] [CrossRef] [PubMed]
- Nakano, C.; Ozawa, H.; Akanuma, G.; Funa, N.; Horinouchi, S. Biosynthesis of aliphatic polyketides by type III polyketide synthase and methyltransferase in Bacillus subtilis. J. Bacteriol. 2009, 191, 4916–4923. [Google Scholar] [CrossRef] [PubMed]
- Funa, N.; Funabashi, M.; Ohnishi, Y.; Horinouchi, S. Biosynthesis of hexahydroxyperylenequinone melanin via oxidative aryl coupling by cytochrome P-450 in Streptomyces griseus. J. Bacteriol. 2005, 187, 8149–8155. [Google Scholar] [CrossRef]
- Song, L.; Barona-Gomez, F.; Corre, C.; Xiang, L.; Udwary, D.W.; Austin, M.B.; Noel, J.P.; Moore, B.S.; Challis, G.L. Type III polyketide synthase β-ketoacyl-ACP starter unit and ethylmalonyl-coA extender unit selectivity discovered by Streptomyces coelicolor genome mining. J. Am. Chem. Soc. 2006, 128, 14754–14755. [Google Scholar] [CrossRef]
- Wenzel, S.C.; Bode, H.B.; Kochems, I.; Müller, R. A type I/type III polyketide synthase hybrid biosynthetic pathway for the structurally unique ansa compound kendomycin. ChemBioChem 2008, 9, 2711–2721. [Google Scholar] [CrossRef]
- Tang, X.; Eitel, K.; Kaysser, L.; Kulik, A.; Grond, S.; Gust, B. A two-step sulfation in antibiotic biosynthesis requires a type III polyketide synthase. Nat. Chem. Biol. 2013, 9, 610–615. [Google Scholar] [CrossRef]
- Aizawa, T.; Kim, S.Y.; Takahashi, S.; Koshita, M.; Tani, M.; Futamura, Y.; Osada, H.; Funa, N. Alkyldihydropyrones, new polyketides synthesized by a type III polyketide synthase from Streptomyces reveromyceticus. J. Antibiot. (Tokyo) 2014, 67, 819–823. [Google Scholar] [CrossRef] [PubMed]
- Čihák, M.; Kameník, Z.; Šmídová, K.; Bergman, N.; Benada, O.; Kofroňová, O.; Petříčková, K.; Bobek, J. Secondary metabolites produced during the germination of Streptomyces coelicolor. Front. Microbiol. 2017, 8, 1–13. [Google Scholar] [CrossRef]
- Petersen, F.; Zähner, H.; Metzger, J.W.; Freund, S.; Hummel, R.P. Germicidin, an autoregulative germination inhibitor of Streptomyces viridochromogenes NRRL B-1551. J. Antibiot. (Tokyo) 1993, 46, 1126–1138. [Google Scholar] [CrossRef] [PubMed]
- Lim, Y.P.; Go, M.K.; Yew, W.S. Exploiting the biosynthetic potential of type III polyketide synthases. Molecules 2016, 21, 1–37. [Google Scholar] [CrossRef] [PubMed]
- Chemler, J.A.; Buchholz, T.J.; Geders, T.W.; Akey, D.L.; Rath, C.M.; Chlipala, G.E.; Smith, J.L.; Sherman, D.H. Biochemical and structural characterization of germicidin synthase: Analysis of a type III polyketide synthase that employs acyl-ACP as a starter unit donor. J. Am. Chem. Soc. 2012, 134, 7359–7366. [Google Scholar] [CrossRef]
- Izumikawa, M.; Shipley, P.R.; Hopke, J.N.; O’Hare, T.; Xiang, L.; Noel, J.P.; Moore, B.S. Expression and characterization of the type III polyketide synthase 1,3,6,8-tetrahydroxynaphthalene synthase from Streptomyces coelicolor A3(2). J. Ind. Microbiol. Biotechnol. 2003, 30, 510–515. [Google Scholar] [CrossRef]
- Chen, H.; Tseng, C.C.; Hubbard, B.K.; Walsh, C.T. Glycopeptide antibiotic biosynthesis: Enzymatic assembly of the dedicated amino acid monomer (S)-3,5-dihydroxyphenylglycine. Proc. Natl. Acad. Sci. USA 2001, 98, 14901–14906. [Google Scholar] [CrossRef] [PubMed]
- Pfeifer, V.; Nicholson, G.J.; Ries, J.; Recktenwald, J.; Schefer, A.B.; Shawky, R.M.; Schröder, J.; Wohlleben, W.; Pelzer, S. A polyketide synthase in glycopeptide biosynthesis: The biosynthesis of the non-proteinogenic amino acid (S)-3,5-dihydroxyphenylglycine. J. Biol. Chem. 2001, 276, 38370–38377. [Google Scholar] [CrossRef] [PubMed]








| Polyketide | Structure | Producer | Type I PKSs | Ref. |
|---|---|---|---|---|
| Avermectin | 16-membered ring macrolide | Streptomyces avermitilis | AVES1-4 | [41] |
| Chalcomycin | 16-membered ring macrolide | Streptomyces bikiniensis | ChmGI-V | [42] |
| Candicidin | 38-membered ring polyene macrolide | Streptomyces griseus IMRU 3570 | CanP1-3, and CanPF | [43,44] |
| FK506 (Tacrolimus) | 23-membered ring macrolide | Streptomyces tsukubaensis, Streptomyces sp. MA6858 | FkbABC | [35,45] |
| FK520 (Ascomycin) | 23-membered ring macrolide | Streptomyces hygroscopicus var. ascomyceticus | FkbABC | [46] |
| Methymycin, Neomethymycin, Narbomycin, Pikromycin | 12-membered ring macrolide, 12-membered ring macrolide, 14-membered ring macrolide, 14- membered ring macrolide | Streptomyces venezuelae | PikAI-IV | [37] |
| Pimaricin | 26-membered ring polyene macrolide | Streptomyces natalensis | PIMS0 and PIMS1 | [47] |
| Rapamycin | 31- membered ring macrolide | Streptomyces hygroscopicus | RAPS1-3 | [34] |
| Spiramycin | 16- membered ring macrolide | Streptomyces ambofaciens | SrmGI-V | [36] |
| Tautomycetin | Linear | Streptomyces sp. CK4412 | TmcA and TmcB | [32] |
| Tylosin | 16- membered ring macrolide | Streptomyces fradiae | TYLGI-V | [48] |
| Polyketide | Intermediate Backbone Structure | Producer | Minimal Type II PKSs | Ref. |
|---|---|---|---|---|
| Medermycin | octaketide | Streptomyces sp. K73 | Med-1,2,23 | [69,70] |
| Doxorubicin | decaketide | Streptomyces peucetius | DpsABCDG | [60,65] |
| Oxytetracycline | decaketide | Streptomyces rimosus | OxyABCD | [71] |
| Gilvocarcin | decaketide | Streptomyces griseoflavus Gö 3592 | GilABC | [72] |
| Oviedomycin | decaketide | Streptomyces antibioticus | OvmPKS | [73] |
| Chartreusin | decaketide | Streptomyces chartreusis | ChaABC | [74] |
| Cervimycin | decaketide | Streptomyces tendae HKI-179 | CerABC | [75,76] |
| Resistomycin | decaketide | Streptomyces resistomycificus | RemABC | [77] |
| Chromomycin | decaketide | Streptomyces griseus subsp. griseus | CmmPKS | [78,79] |
| Hedamycin | dodecaketide | Streptomyces griseoruber | HedCDE | [80,81] |
| Fredericamycin | pentadecaketide | Streptomyces griseus ATCC 49344 | FdmFSGH | [82,83] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Risdian, C.; Mozef, T.; Wink, J. Biosynthesis of Polyketides in Streptomyces. Microorganisms 2019, 7, 124. https://0-doi-org.brum.beds.ac.uk/10.3390/microorganisms7050124
Risdian C, Mozef T, Wink J. Biosynthesis of Polyketides in Streptomyces. Microorganisms. 2019; 7(5):124. https://0-doi-org.brum.beds.ac.uk/10.3390/microorganisms7050124
Chicago/Turabian StyleRisdian, Chandra, Tjandrawati Mozef, and Joachim Wink. 2019. "Biosynthesis of Polyketides in Streptomyces" Microorganisms 7, no. 5: 124. https://0-doi-org.brum.beds.ac.uk/10.3390/microorganisms7050124