1. Introduction
One of the highest public health challenges worldwide is the increase in the number and types of antimicrobial resistances (AMR) [
1]. The use and misuse of antimicrobials in human medicine is one of the main causes of this increasing problem, but inappropriate practices in intensive livestock farms have also contributed to the alarming increase in antibiotic resistant bacteria (ARB) [
2,
3]. Antibiotics used in animal production with different purposes (therapeutically and prophylactically) are finally disseminated through the environment. It has been estimated that about 75% of the administered antibiotics is not absorbed by animals and is excreted via the feces or urine [
4]. In this sense, García-Galán et al., 2011 [
5] reported the presence of emerging pollutants in the Ebro basin (area with intensive livestock farms), including at least eight types of antibiotics. As a consequence of the antibiotic pressure, ARB have been isolated from different sources such as farms (manure), water and meat products [
4,
6,
7,
8]. The dissemination of AMR throughout the environment represents a risk to human health [
9,
10]. In particular, one of the main routes for the dissemination of ARB and resistance genes (ARGs) is the aquatic environment [
11,
12]. Therefore, a One Health approach is needed to address the problem of antimicrobial resistance. Recently, the World Health Organization (WHO) has published a list of antibiotic-resistant priority pathogens with the aim to promote research and development of new antibiotics, as one of the proposed strategies to control the problem of global resistance to antimicrobial medicines [
13]. The list includes 12 families of bacteria that pose the greatest threat to human health, especially if they are spread throughout the environment. Among pathogens classified as priority 1 (critical) and 2 (high), carbapenem, β-lactam, vancomycin and methicillin resistances are considered. β-lactam antibiotics have been the most extended therapeutic choice for the treatment of human and animal infections worldwide, and consequently, bacteria have developed different β-lactam resistance mechanisms, such as the production of extended spectrum β-lactamases (ESBLs) and carbapenemases [
14]. The presence of
bla genes encoding SHV, TEM, CTX-M groups, KPC, NDM and VIM enzymes has been frequently reported in rivers of different regions over the world [
15,
16]. Despite methicillin-resistant
Staphylococcus aureus (MRSA) has been detected basically in clinical environments [
17,
18], the presence of MRSA
mecA/C in river water has been described [
19]. Regarding vancomycin resistant enterococci (VRE), although the presence of these bacteria seems to be related to small wild mammals, rabbits and birds [
20,
21], some authors described the presence of
E. faecium vanA and
vanB in wastewater and surface waters [
22,
23]. Colistin has become as the last alternative in human medicine for the treatment of infections due to multidrug-resistant Gram-negative bacteria [
24]. Colistin sulfate is used for the control of Enterobacteriaceae infections in pig production in some countries [
25,
26], contributing to the spread of colistin resistances mediated by the transferable plasmid
mcr-1 [
27]. In this context, it would be very interesting to search for this type of resistance in different aquatic environments, such as rivers, WWTPs and collectors.
POCTEFA 2014-2020 is the acronym for the INTERREG V-A Spain-France-Andorra Program (
https://www.poctefa.eu/). It is a European territorial cooperation program created to promote the sustainable development of the regions near to the Franco-Spanish border (Navarra, Huesca, Zaragoza, Lleida, Pyrénées-Atlantiques, Hautes-Pyrénées, Orientals-Pyrénées, Haute Garonne and Ariege). This area covers a region of 115.583 km
2, populated by 15 million habitants, being the intensive livestock farms as the main rural economic engine (especially porcine, poultry and rabbit production). In this sense, the main objective of this study was to determine the presence of ARB in rivers, wastewater treatment plants (WWTPs) and collectors in the North of Spain and South of France (hereafter named POCTEFA area). Specifically, we focused the study on the isolation and characterization of critical and high priority resistant pathogens according to the WHO list: Enterobacteriaceae,
Pseudomonas aeruginosa and
Acinetobacter baumanii carbapenem-resistant; Enterobacteriaceae ESBL-producing;
Enterococcus faecium vancomycin-resistant and
Staphylococcus aureus methicillin-resistant. In addition, due to the recent interest in colistin resistances, we also included the search for Enterobacteriaceae colistin-resistant.
2. Materials and Methods
2.1. Sample Collection
The sampling was performed by trained people from the University of Zaragoza (Laboratory of Water and Environmental Health) in 40 locations from POCTEFA area including rivers, WWTPs and collectors (hospital and slaughterhouses). A total of 80 samples were collected in two seasons of 2018 (April–May and October–November). Rivers were located in France and Spain, whereas WWTPs and collectors were present in Navarra (Northern Spain;
Figure 1). Sampling of WWTPs was performed in influent and effluent water and sampling of rivers was done upstream and downstream of the WWTP (when present). Complete information of each point, provided by the Laboratory of Water and Environmental Health of University of Zaragoza, is available in the
supplementary material (Tables S1 and S2).
Samples were taken in sterile containers in accordance with ISO 19458 [
28] and ISO 5667-3 [
29] standards and stored at 5 ± 3 °C in the dark until they were sent to the University of Navarra. Microbiological analysis was carried out within 24 h of arrival of samples (stored at 5 ± 3 °C).
2.2. Isolation and Identification of Resistant Bacteria
In order to detect the presence of resistant bacteria (even the lethargic ones by environmental stressors such as the temperature or the lack of nutrients), two approaches were performed. In the first method, 1 mL of each sample was spread on the surface of specific selective culture media for each resistance type (described below). In the second method, two previous enrichment processes were carried out for the recovery of stressed cells. This way, 10 mL water samples were transferred to 10 mL of double concentration EE Mossel broth (Difco, Le Pont de Claix, France) and were incubated at 37 ± 1 °C during 24 h, in order to isolate Gram negative bacteria. Similarly, enrichment in Giolitti Cantoni broth (Oxoid, Basingstoke, United Kingdom) was performed for the recovery of Gram positive bacteria (24–48 h at 37 ± 1 °C). Following the incubation periods, isolations were performed on the selected culture media. In the case of carbapenem and colistin resistances, selective culture media was changed in the second sampling in order to improve the recovery of these strains (taking into account the obtained results in the first sampling).
Chromogenic selective plates from bioMerieux (Marcy l’Etoile, France) were used for the isolation of the target resistant bacteria. Thus, ChromID ESBL plates containing a mixture of antibiotics including cefpodoxime (CPD) were used for the isolation of suspicious ESBL-producing strains. ChromID MRSA contains cefoxitin (FOX) as a selective agent and was used for the isolation of MRSA. ChromID VRE agar plates selects vancomycin (VA) resistant Enterococcus, allowing the differentiation between E. faecium and E. faecalis. Finally, ChromID CARBA SMART agar plates and ChromID CARBA agar plates were used for the isolation of carbapenemase-producing Enterobacteriaceae (CPE; first and second sampling events, respectively). In addition, Columbia CNA and MacConkey agar supplemented with 2 µg/mL of colistin (COL; Oxoid) were used for the isolation of colistin resistant bacteria (first and second sampling events, respectively). After the incubation at 37 ± 1 °C during 24–48 h, suspicious colonies were isolated on ChromID CPSE, nutrient agar or blood agar (bioMerieux). The identification was carried out using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS; bioMerieux) or biochemical tests (oxidase, API 20NE, API20E, APIstaph or API 20STREP; bioMerieux). Pure cultures were stored at −80 °C for further characterization.
2.3. Phenotypic and Genotypic Characterization of Resistant Strains
The antibiotic disks used for the phenotypic characterization were provided by Becton Dickinson (Le Pont de Claix, France), ROSCO Diagnostica (Taastrup, Denmark) and by Biomerieux in the case of the E-tests. The results were interpreted according to “Clinical & Laboratory Standars Institute”, CLSI [
30] or “The European Committee on Antimicrobial Susceptibility Testing”, EUCAST guidelines [
31]. The antimicrobials tested and resistance breakpoints can be found in the
supplementary material (Table S3). The specific methodology applied for the phenotypic and/or genotypic characterization of each types of resistance is explained in the subsequent sections.
2.3.1. ESBL-Producer Enterobacteriaceae and Other β-Lactamases
ESBL production was confirmed by the double-disk synergy test (DDST) according to Jarlier et al., 1988 [
32]. Basically, the amoxicillin/clavulanic acid (AMC, 30 μg) was placed in the center of the inoculated Mueller Hinton cation-adjusted agar plate (MH; Becton Dickinson) and the following β-lactam antibiotics were placed at a distance of 20 mm: ceftazidime (CAZ, 30 μg), ceftriaxone (CRO, 30 μg), aztreonam (AZT, 30 μg) and cefpodoxime (CPD, 10 μg). After incubation at 37 ± 1 °C for 18–24 h, the strain was considered as the ESBL-producer when the enhanced inhibition zone was observed between the cephalosporin disk and AMC, indicating synergy. AmpC β-lactamase production was determined following the methodology of Thean et al., 2009 [
33] by comparing the diameters of each β-lactam or β-lactam with an inhibitor (ceftazidime/clavulanic acid and cefotaxime/clavulanic acid) in MH and MH supplemented with cloxacillin (250 mg/L, Sigma Aldrich, Singapore). When an increased inhibition zone of >5 mm in cloxacillin plates was observed, the microorganisms was considered to be an AmpC-producer [
34]. Finally, we studied the presence of metallo-β-lactamases (MBL) according to Arakawa et al., 2000 [
35], using CAZ (30 μg), imipenem (IMP, 10 μg) and EDTA (10 μL) disks in MH plates. In addition, an IMP disk was used to which 10 µL of EDTA was added. It was considered an MBL-producing strain when a synergistic effect was observed between the IMP, CAZ and EDTA discs and if the difference between the IMP + EDTA disc and the IMP disc was >5 mm.
The DNA extraction procedure was performed with the DNeasy® Blood and Tissue kit (Qiagen, Barcelona, Spain), using a pretreatment protocol for Gram-negative bacteria and following the manufacturer’s instruction. The quantity and quality of the DNA was analyzed using a Nanodrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA).
The detection of AmpC β-lactamases genes was performed using the multiplex-PCR assay described by Pérez-Pérez and Hanson, 2002 [
36]. The primers, size of the amplicons and conditions followed are summarized in
Table 1.
The identification of
blaTEM,
blaSHV and
blaOXA genes was performed using the multiplex-PCR assay described by Colom et al., 2003 [
37] while a modification of the multiplex-PCR described by Woodford et al., 2006 [
38] was used for the study of
blaCTX-M genes. The reaction mixture composition and amplification conditions for the
blaCTX-M genes were described in the manuscript of Ojer-Usoz et al., 2014 [
8]. All the details for the several multiplex PCR assays are shown in
Table 1.
A bidirectional DNA sequence analysis of the amplicons were performed by the Macrogen EZ-Seq purification service to determine the molecular types of
bla genes (Macrogen Europe, Amsterdam, The Netherlands). Searches for DNA and protein homologies were carried out through the National Centre for Biotechnology Information (
http://0-www-ncbi-nlm-nih-gov.brum.beds.ac.uk/) using the BLAST program. The alignment of DNA and amino acids sequences was performed using Clustal Omega (
http://www.ebi.ac.uk/Tools/msa/clustalo/).
2.3.2. Carbapenemase-Producing Strains
Disks of ertapenem (ETP, 10 μg, Oxoid), IMP (10 μg, Oxoid) and meropenem (MER, 10 μg, Becton Dickinson) were used to determine carbapenemase production. In addition to the screening of the presence of β-lactamase and metalo-β-lactamase (described above), OXA-48-like, KPC, NDM and VIM type carbapenemases were determined using the immunochromatography test Resist-4 O.K.N (Coris Bioconcept, Gembloux, Belgium), according to the manufacturer instructions.
2.3.3. Colistin Resistant Enterobacteriaceae
COL E-test (Biomerieux) was performed for the determination of this resistance, using EUCAST guidelines [
31] for the interpretation of the inhibition zone (
Table S3). The presence of
mcr-1,
mcr-2,
mcr-3,
mcr-4 and
mcr-5 genes was detected by conventional PCRs using the specific primers and conditions shown in
Table 2 and following the conditions described in the corresponding works [
39,
40,
41,
42,
43].
2.3.4. Methicillin Resistant Strains
Methicillin resistances were confirmed by using FOX disks (30 μg, Becton Dickinson). For the determination of gene mecA, the AlereTM PBP2a test was performed according to the manufacturer instructions (Abbot, Scarborough, Maine). This is a rapid qualitative immunochromatographic analysis for the detection of penicillin 2a binding protein (encoded by mecA).
2.3.5. Vancomycin Resistant Strains
The VA resistance of
E. faecium was confirmed by the E-test (bioMerieux), in Mueller Hinton Agar with 5% sheep blood (bioMerieux). Additional E-test of teicoplanin (TEC; bioMerieux) was performed in positive strains in order to determine the presence of gene
vanA or
vanB. Strains with the
vanA phenotype are characterized by a high level of resistance to both VA (MIC ≥ 64 µg/mL) and TEC (MIC ≥ 16 µg/mL). However, strains carrying the
vanB gene are characterized by variable levels of resistance to VA (MIC between 4 and ≥ 1000 µg/mL) and sensitivity to TEC [
20].
2.4. Antimicrobial Resistance Patterns
The antimicrobial susceptibility of resistant strains to additional antibiotics was obtained in the MicroScan® system (Siemens AG, Munich, Germany). NM37, PN28 and Neg Multidrug Resistant MIC 1 panels (Siemens AG, Germany) were used in combination with Lab Pro® 3.5 software for determining the minimum inhibitory concentrations (MICs). The panels included the following antimicrobials: AMC, ampicillin (AMP), ampicillin-sulbactam (AMS), azithromycin (AZI), AZT, cefazolin (CZ), cefepime (FEP), CAZ, cefuroxime (CXM), CPD, cefotaxime (CTX), FOX, chloramphenicol (CHL), ciprofloxacin (CIP), COL, clindamycin (Cd), daptomycin (DAP), ETP, erythromycin (ERY), fosfomycin (FOT), fusidic acid (FA), gentamicin (GM), IMP, levofloxacin (LV), linezolid (Lz), MER, mupirocin (MUP), moxifloxacin (MXF), mezlocillin (MZ), norfloxacin (NOR), nitrofurantoin (FD), oxacillin (OX), penicillin (P), piperacillin (PIP), piperacillin-tazobactam (TZP), rifampicin (RA), synercid (SYN), tobramycin (TO), tetracycline (TET), tigecycline (TIG), TEC, trimethoprim-sulfamethoxazole (SXT) and VA.
This automated method provided very interesting results for the study of ESBL-producing bacteria. ESBL production was confirmed when a > 3 two-fold concentration decrease occurred in an MIC for any of β-lactams tested in combination with clavulanic acid versus its MIC when tested alone [
8,
44]. The MIC50 and MIC90 (minimum concentration required to inhibit the growth of 50% and 90% of organisms, respectively) were used to evaluate antibiotic sensitivities. Multi-drug resistances (MDR) and extensive MDR were considered when resistances to three or at least five antimicrobial agents were detected, respectively [
45].
2.5. Statistical Analysis
The results for the rates of resistances to antibiotics were subjected to statistical processing with the SPSS 15 software (SPSS Inc., Chicago, IL, USA), applying the Chi-square (X2) test with a level of significance of p < 0.05.
4. Discussion
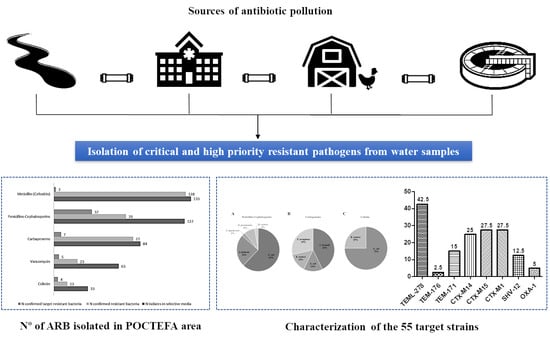
This study aimed to determine the prevalence of antibiotic resistant bacteria in aquatic environments of the POCTEFA area, a region of intensive livestock activity. The widespread presence of resistant bacteria observed in rivers, WWTPs and collectors (96.4% and 100%, respectively), highlight the impact of human activity on the spread of these resistances, especially from hospital and livestock production. In fact, 55 resistant strains identified as critical and high priority resistant pathogens (according to WHO list) were isolated in the study.
The wastewater from slaughterhouses is considered a relevant source of antimicrobial resistant bacteria and consequently may be important for its diffusion into the environment [
46]. The livestock pressure in most of the Spanish rivers studied in this work was high, where pig farms stood out mainly (
Table S1). This could be the reason of the higher prevalence of resistances in the Spanish rivers than the French ones (
Table 3). In addition, the unique negative river regarding the presence of resistant bacteria (9_ASE) was located in the Pyrenees area, where no relevant cattle exploitations such as pigs, birds and rabbits were reported.
In agreement with our results, previous studies reported an increase of ESBL and carbapenemase-producing Enterobacteriaceae (CPE) in rivers and WWTPs, with high percentages of clinically relevant multidrug resistant bacteria and related genes (
intI1,
sul1,
blaOXA,
mcr-1,
blaCTX-M15,
blaKPC and
blaVIM, among others) that were still present in effluent samples, indicating an insufficient reduction during conventional wastewater treatment [
47]. In this line, our results are in agreement with previous work published by Ojer-Usoz in the same region of Navarra [
8], with a similar prevalence of ESBL after 6 years. In addition, the association between
blaOXA-1 and resistance to aminoglycosides and quinolones reported by Osińska et al., 2016 [
48] was confirmed in the present study, because the two WWTP isolates carrying
blaOXA-1 were resistant to TO, LV, CIP, MXF and NOR. Indeed, the increased quinolone resistance rate in the isolates (50%) may be caused by the use of enrofloxacin in slaughtered broiler herds [
46]. Furthermore, the high prevalence regarding AmpC β-lactamases could be related with the large number of
Serratia and
Citrobacter strains (carriers of chromosomal AmpC). In addition, it is interesting to keep in mind that ESBL and AmpC coproduction was detected in 19 strains, despite only five strains being confirmed as AmpC-producers by molecular methods. Moreover, two of them (numbers 39 and 42 in
Table 5), were also carriers of carbapenemase gene KPC. The isolation of different KPC producing species (
E. coli,
K. oxytoca and
C. freundii) in the same water sample (33_ARD3e) reinforced the hypothesis that a horizontal gene transfer is taking place between different bacterial species. Finally, in this study we did not isolate
A. baumanii resistant to penicillins, cephalosporins or carbapenems. This could be related to the low presence of this pathogen or with methodological problems on the isolation of this species.
Colistin (polymyxin E) is currently used as a last alternative drug against MDR Gram negative bacteria. However, its resistance has even emerged in humans since it has been widely used in pig production and in some countries in veterinary (especially in cows) for the treatment of gastrointestinal infections caused by Enterobacteriaceae [
26]. This resistance is frequently related to chromosomal mutations, nonetheless, the mechanism by which the
mcr-1 gene confers resistance to COL was the first one that described plasmid mediated transmission antibiotic resistance and was first discovered in China on a pig farm [
39]. Despite this gene being widespread in the environment [
49] and it having been documented in 30 strains isolated from three Spanish WWTPs [
50], only one
E. coli strain isolated from a rabbit slaughterhouse collector (number 47 in
Table 5) was positive for
mcr-1 in our study.
Enterococci are recognized as important nosocomial pathogens due to their natural intrinsic resistance and their ability to acquire resistance to multiple drugs [
49]. The resistance to VA in enterococci (VRE) is associated with the use of this antibiotic in clinics and, as a consequence, effluents from hospitals constitute an important point for the transmission of this resistance [
51]. In this sense, despite the fact that VA resistant bacteria were isolated from the hospital collector (37_ARH), none of these isolates was identified as VRE. However, VRE were present in the influent waters of the WWTPs of two points near hospitals (29_ARD1e and 33_ARD3e), in accordance with other studies [
52]. In general, VA resistances are specially linked to
vanA and
vanB genes and represent a major public health problem, due to their resistant gene transfer capacity [
53]. In this sense,
vanB carriers are characterized by high levels of VA resistance and TEC sensitivity, and the resistance is transferred by conjugation associated with the mobilization of genetic material through the acquisition and/or exchange of transposons [
20]. In agreement with that, all our VRE isolates showed the
vanB phenotype. As VA is not used in veterinary medicine, the use of other glycopeptides as an animal growth promoter (such as avoparcin), was associated with the increase in VRE in the 70s [
20]. Numerous studies have shown that VRE persisted in animals for a long time after avoparcin was banned [
49]. Therefore, the presence of identical resistance genes in animal and human enterococci, suggest the spread between isolates from different environments [
53]. The isolation of
E. faecium VA resistance in samples from a rabbit slaughterhouse collector (40_ARM) reinforces this hypothesis. Finally, it is known that VRE can rapidly develop resistance after the introduction of new antimicrobial agents in the clinic, such as quinupristin-dalfopristin (SYN), Lz and DAP [
54]. So, it should be noted that 80% of the
E. faecium vanB isolates of this study were resistant to SYN and 40% were resistant to DAP, whereas no resistances to Lz were observed.
One of the most important acquired resistances in
S. aureus is methicillin resistance (MRSA) and is mainly due to the acquisition of the
mecA gene, encoding a β-lactam low affinity penicillin binding protein (PBP) called PBP2a [
49]. In general MRSA isolates from surface water are quite rare, with only a low number of isolates [
51]. Despite this, MRSA
mecA has been reported to survive in rivers and municipal wastewater and had been associated with colonized people [
19]. In addition, the presence of gene
mecC has been reported for the first time in a Spanish river, highlighting the potential role of water in the dissemination of
mecC MRSA [
19].
S. aureus mecC was also isolated from animals and an urban wastewater treatment plant [
55] and other studies highlighted the emergence of
S. aureus mecC in livestock production, particularly in pigs in European countries [
56]. In this sense, our two MRSA strains (negative for the
mecA gene in the PBP2a test) were isolated from rivers (numbers 54 and 55 in
Table 6) with high incidence from pig exploitations, which would reinforce this hypothesis.
In general, the main objective of wastewater treatment is to eliminate organic (chemical and biological) components, phosphorous and nitrogen nutrients as well as suspended solids. Directive 91/271/EEC [
57] establishes the guidelines to be followed by the Member States of the European Union to ensure that urban wastewater receives adequate treatment before discharge, but it does not include disinfection processes that reduce the microbiological charge and ARGs in the effluents [
58]. Consequently, these bacteria are incorporated into the environment through the direct or indirect discharge of treated water or through sludge, which finally is used as a fertilizer in agricultural practices. In the same way, the directive does not provide specific restrictions for effluents from hospital wastewater, which also constitute an important reservoir of ARB [
52]. It is known that some ARB can be removed through conventional wastewater treatment processes [
6], but still large numbers that survive in the effluent. Therefore, tertiary treatment methods or advanced treatment technologies are those that manage to eliminate some bacterial load and genes [
47]. In this sense, UV and ozone-treatment have been investigated for a long time with the aim of reducing these microbial loads. UV disinfection contributes to the effective reduction of some bacteria, like 99.9% of MRSA or VRE [
6]. However, Munir et al., 2011 [
59] founded that this disinfection did not contribute to the reduction of TET and sulfonamide resistant bacteria. Moreover, ozonation is an efficient process to eliminate organic microcontaminants and for inactivating bacteria through the production of highly reactive radical [
60]. Other tertiary treatments are based on the water exposure to solar radiation in the lagoon and according to López Martínez [
61] are able to reduce the microbiological concentration up to four orders of magnitude at the longest time of exposure to solar radiation. However, these advanced wastewater treatment technologies are also known to accelerate horizontal gene transfer due to the activation of different repair mechanisms involved in the dissemination of antibiotic resistance genes [
6]. Consequently, it is necessary to develop other additional strategies and guidelines for the elimination of microbial contaminants in wastewater, which included surveillance of pathogenic bacteria and ARGs. For that reason, there is a need to improve effective disinfection measures and treatments in WWPTs and animal slaughterhouses to avoid environmental contamination and prevent the evolution of antibiotic resistance.