Targeting Cancer Stem Cells with Novel 4-(4-Substituted phenyl)-5-(3,4,5-trimethoxy/3,4-dimethoxy)-benzoyl-3,4-dihydropyrimidine-2(1H)-one/thiones
Abstract
:1. Introduction
2. Results and Discussion
3. Material and Methods
3.1. Experimental
3.2. General Synthesis of 4-(Substituted phenyl)-5-(3,4,5-trimethoxybenzoyl/3,4-dimethoxybenzoyl)-3,4-dihydropyrimidin-2(1H)-ones DHP 1–9
3.3. Cell line and Tissue Culture
3.4. Flow Cytometric Analysis of Cellular DNA Content
3.5. Side Population Staining by DYECYCLE Violet Stain
3.6. Antitumor Activity in Mice
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Winquist, R.J.; Furey, B.F.; Boucher, D.M. Cancer stem cells as the relevant biomass for drug discovery. Curr. Opin. Pharmacol. 2010, 10, 385–390. [Google Scholar] [CrossRef] [PubMed]
- McDermott, S.P.; Wicha, M.S. Targeting breast cancer stem cells. Mol. Oncol. 2010, 4, 404–419. [Google Scholar] [CrossRef] [PubMed]
- Bonnet, D.; Dick, J.E. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat. Med. 1997, 3, 730–737. [Google Scholar] [CrossRef] [PubMed]
- Al-Hajj, M.; Wicha, M.S.; Benito-Hernandez, A.; Morrison, S.J.; Clarke, M.F. Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. USA 2003, 100, 3983–3988. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.K.; Clarke, I.D.; Terasaki, M.; Bonn, V.E.; Hawkins, C.; Squire, J.; Dirks, P.B. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003, 63, 5821–5828. [Google Scholar] [PubMed]
- Ho, M.M.; Ng, A.V.; Lam, S.; Hung, J.Y. Side population in human lung cancer cell lines and tumors is enriched with stem-like cancer cells. Cancer Res. 2007, 67, 4827–4833. [Google Scholar] [CrossRef] [PubMed]
- Ricci-Vitiani, L.; Lombardi, D.G.; Pilozzi, E.; Biffoni, M.; Todaro, M.; Peschle, C.; de Maria, R. Identification and expansion of human colon-cancer-initiating cells. Nature 2007, 445, 111–115. [Google Scholar] [CrossRef] [PubMed]
- Bomken, S.; Fiser, K.; Heidenreich, O.; Vormoor, J. Understanding the cancer stem cell. Br. J. Cancer 2010, 103, 439–445. [Google Scholar] [CrossRef] [PubMed]
- Ginestier, C.; Hur, M.H.; Charafe-Jauffret, E.; Monville, F.; Dutcher, J.; Brown, M.; Jacquemier, J.; Viens, P.; Kleer, C.G.; Liu, S.; et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell 2007, 1, 555–567. [Google Scholar] [CrossRef] [PubMed]
- Morimoto, K.; Kim, S.J.; Tanei, T.; Shimazu, K.; Tanji, Y.; Taguchi, T.; Tamaki, Y.; Terada, N.; Noguchi, S. Stem cell marker aldehyde dehydrogenase 1-positive breast cancers are characterized by negative estrogen receptor, positive human epidermal growth factor receptor type 2, and high Ki67 expression. Cancer Sci. 2009, 100, 1062–1068. [Google Scholar] [CrossRef] [PubMed]
- Charafe-Jauffret, E.; Ginestier, C.; Iovino, F.; Tarpin, C.; Diebel, M.; Esterni, B.; Houvenaeghel, G.; Extra, J.M.; Bertucci, F.; Jacquemier, J.; et al. Aldehyde dehydrogenase 1-positive cancer stem cells mediate metastasis and poor clinical outcome in inflammatory breast cancer. Clin. Cancer Res. 2010, 16, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Tanei, T.; Morimoto, K.; Shimazu, K.; Kim, S.J.; Tanji, Y.; Taguchi, T.; Tamaki, Y.; Noguchi, S. Association of breast cancer stem cells identified by aldehyde dehydrogenase 1 expression with resistance to sequential Paclitaxel and epirubicin-based chemotherapy for breast cancers. Clin. Cancer Res. 2009, 15, 4234–4241. [Google Scholar] [CrossRef] [PubMed]
- Kondo, T.; Setoguchi, T.; Taga, T. Persistence of a small subpopulation of cancer stem-like cells in the C6 glioma cell line. Proc. Natl. Acad. Sci. USA 2004, 101, 781–786. [Google Scholar] [CrossRef] [PubMed]
- Patrawala, L.; Calhoun, T.; Schneider-Broussard, R.; Zhou, J.; Claypool, K.; Tang, D.G. Side population is enriched in tumorigenic, stem-like cancer cells, whereas ABCG2+ and ABCG2− cancer cells are similarly tumorigenic. Cancer Res. 2005, 65, 6207–6219. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Lewis, M.T.; Huang, J.; Gutierrez, C.; Osborne, C.K.; Wu, M.F.; Hilsenbeck, S.G.; Pavlick, A.; Zhang, X.; Chamness, G.C.; et al. Intrinsic resistance of tumorigenic breast cancer cells to chemotherapy. J. Natl. Cancer Inst. 2008, 100, 672–679. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Yao, H.; Zhu, P.; Zhang, X.; Pan, Q.; Gong, C.; Huang, Y.; Hu, X.; Su, F.; Lieberman, J.; et al. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell 2007, 131, 1109–1123. [Google Scholar] [CrossRef] [PubMed]
- Al-Hajj, M.; Becker, M.W.; Wicha, M.; Weissman, I.; Clarke, M.F. Therapeutic implications of cancer stem cells. Curr. Opin. Genet. Dev. 2004, 14, 43–47. [Google Scholar] [CrossRef] [PubMed]
- Bokaeva, S.S. The effects of some pyrimidine derivates on the growth of transplanted tumors in animals. Tr. Kaz. Nauchno-Issled. Inst. Onkol. Radiol. 1967, 3, 305–309. [Google Scholar]
- Mayer, T.U.; Kapoor, T.M.; Haggarty, S.J.; King, R.W.; Schreiber, S.L.; Mitchison, T.J. Small molecule inhibitor of mitotic spindle bipolarity identified in a phenotype-based screen. Science 1999, 286, 971–974. [Google Scholar] [CrossRef] [PubMed]
- Russowsky, D.; Canto, R.F.; Sanches, S.A.; D’Oca, M.G.; de Fátima, A.; Pilli, R.A.; Kohn, L.K.; Antônio, M.A.; de Carvalho, J.E. Synthesis and differential antiproliferative activity of Biginelli compounds against cancer cell lines: Monastrol, oxo-monastrol and oxygenated analogues. Bioorg. Chem. 2006, 34, 173–182. [Google Scholar] [CrossRef] [PubMed]
- Prokopcova, H.; Dallinger, D.; Uray, G.; Kaan, H.Y.; Ulaganathan, V.; Kozielski, F.; Laggner, C.; Kappe, C.O. Structure-activity relationships and molecular docking of novel dihydropyrimidine-based mitotic Eg5 inhibitors. ChemMedChem 2010, 5, 1760–1769. [Google Scholar] [CrossRef] [PubMed]
- Wright, C.M.; Chovatiya, R.J.; Jameson, N.E.; Turner, D.M.; Zhu, G.; Werner, S.; Huryn, D.M.; Pipas, J.M.; Day, B.W.; Wipf, P.; et al. Pyrimidinone-peptoid hybrid molecules with distinct effects on molecular chaperone function and cell proliferation. Bioorg. Med. Chem. 2008, 16, 3291–3301. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Saez, I.; DeBonis, S.; Lopez, R.; Trucco, F.; Rousseau, B.; Thuéry, P.; Kozielski, F. Structure of human Eg5 in complex with a new monastrol-based inhibitor bound in the R configuration. J. Biol. Chem. 2007, 282, 9740–9747. [Google Scholar] [CrossRef] [PubMed]
- Ramos, L.M.; Guido, B.C.; Nobrega, C.C.; Corrêa, J.R.; Silva, R.G.; de Oliveira, H.C.; Gomes, A.F.; Gozzo, F.C.; Neto, B.A. The biginelli reaction with an imidazolium-tagged recyclable iron catalyst: Kinetics, mechanism, and antitumoral activity. Chem. Eur. J. 2013, 19, 4156–4168. [Google Scholar] [CrossRef] [PubMed]
- Soumyanarayanan, U.; Bhat, V.G.; Kar, S.S.; Mathew, J.A. Monastrol mimic Biginelli dihydropyrimidinone derivatives: Synthesis, cytotoxicity screening against HepG2 and HeLa cell lines and molecular modeling study. Org. Med. Chem. Lett. 2012, 2. [Google Scholar] [CrossRef] [PubMed]
- Kumar, B.R.P.; Sankar, G.; Nasir Baig, R.B.; Chandrashekaran, S. Novel Biginelli dihydropyrimidines with potential anticancer activity: A parallel synthesis and CoMSIA study. Eur. J. Med. Chem. 2009, 44, 4192–4198. [Google Scholar] [CrossRef] [PubMed]
- Sample Availability: Samples of the compounds (DHP 1–9) in pure form are available from the authors.







| Compounds | R | R1 | R2 | (Yield %) | m.p. (°C) |
|---|---|---|---|---|---|
| DHP-1 | Phenyl | OCH3 | O | 70 | 153–155 |
| DHP–2 | 4-Chlorophenyl | OCH3 | O | 75 | 138–140 |
| DHP-3 | 4-Nitrophenyl | OCH3 | O | 65 | 158–160 |
| DHP-4 | 3,4-Dimethoxyphenyl | OCH3 | O | 72 | 165–167 |
| DHP-5 | 4-Ethoxyphenyl | OCH3 | O | 60 | 168–170 |
| DHP-6 | Phenyl | H | S | 65 | 248–250 |
| DHP-7 | 4-Chlorophenyl | H | S | 65 | 243–245 |
| DHP-8 | 4-Nitrophenyl | H | S | 68 | 258–260 |
| DHP-9 | 3,4-Dimethoxyphenyl | H | S | 70 | 228–230 |
| Compounds | * Side Population (%) at 10 μM | # Side Population Inhibition (%) at 10 μM |
|---|---|---|
| DHP-1 | 4.90 ± 0.2 | 0 |
| DHP-2 | 1.72 ± 0.1 | 64.7 |
| DHP-3 | 1.76 ± 0.3 | 64 |
| DHP-4 | 1.44 ± 0.5 | 70.5 |
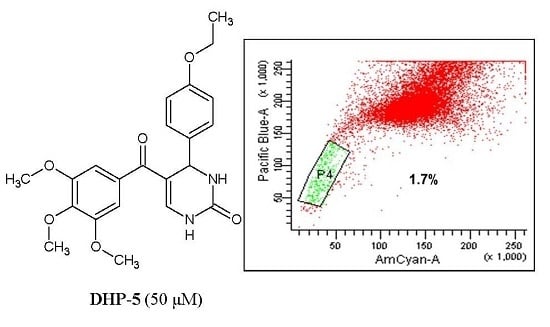
| DHP-5 | 2.01 ± 0.7 | 58.82 |
| DHP-6 | 1.47 ± 0.6 | 70 |
| DHP-7 | 4.90 ± 0.3 | 0 |
| DHP-8 | 2.4 ± 0.1 | 50 |
| DHP-9 | 4.90 ± 0.1 | 0 |
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bhat, M.A.; Al-Dhfyan, A.; Al-Omar, M.A. Targeting Cancer Stem Cells with Novel 4-(4-Substituted phenyl)-5-(3,4,5-trimethoxy/3,4-dimethoxy)-benzoyl-3,4-dihydropyrimidine-2(1H)-one/thiones. Molecules 2016, 21, 1746. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules21121746
Bhat MA, Al-Dhfyan A, Al-Omar MA. Targeting Cancer Stem Cells with Novel 4-(4-Substituted phenyl)-5-(3,4,5-trimethoxy/3,4-dimethoxy)-benzoyl-3,4-dihydropyrimidine-2(1H)-one/thiones. Molecules. 2016; 21(12):1746. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules21121746
Chicago/Turabian StyleBhat, Mashooq Ahmad, Abdullah Al-Dhfyan, and Mohamed A. Al-Omar. 2016. "Targeting Cancer Stem Cells with Novel 4-(4-Substituted phenyl)-5-(3,4,5-trimethoxy/3,4-dimethoxy)-benzoyl-3,4-dihydropyrimidine-2(1H)-one/thiones" Molecules 21, no. 12: 1746. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules21121746