Octadecaneuropeptide (ODN) Induces N2a Cells Differentiation through a PKA/PLC/PKC/MEK/ERK-Dependent Pathway: Incidence on Peroxisome, Mitochondria, and Lipid Profiles
Abstract
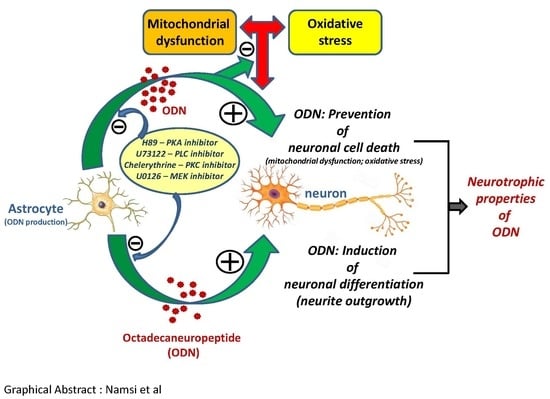
:1. Introduction
2. Materials and Methods
2.1. Cell Culture and Treatments
2.2. Evaluation of Neuronal Differentiation With Morphological Criteria
2.3. Fluorimetric Measurement of Cell Viability with the FDA Assay
2.4. Flow Cytometric Measurement of Transmembrane Mitochondrial Potential with DiOC6(3)
2.5. Visualization of Neurite Formation by Staining with Sulforhodamine 101
2.6. Simultaneous Observation of Mitochondria and Peroxisomes by Fluorescence Microscopy: Detection of Mitochondria after Staining with MitoTracker Red and of Peroxisomes by Indirect Immunofluorescence with an Antibody Directed Against the ABCD3 Peroxisomal Transporter
2.7. Transmission Electron Microscopy of Mitochondria and Peroxisomes
2.8. Evaluation of the Activation of PKA, PLC, PKC and MEK/ERK Signaling Pathways in ODN-Treated N2a Cells
2.9. Determination of the Fatty Acid Profile by Gas Chromatography
2.10. Isotope Dilution Mass Spectrometry Analysis for Cholesterol and Cholesterol Precursors
2.11. Statistical Analysis
3. Results
3.1. Quantification of Neuronal Differentiation of N2a Cells Induced by ODN
3.2. Characterization of N2a Differentiated Cells after Coloration with Sulforhodamine 101
3.3. Effect of ODN on the Topography of Mitochondria and Peroxisomes in N2a Differentiated Cells: Evaluation by Fluorescence and Transmission Electron Microscopy
3.4. Characterization with Different Inhibitors of the Signaling Pathways Involved in the Neuronal Differentiation of ODN-Treated N2a Cells
3.5. Effect of ODN on Fatty Acid, Cholesterol and Cholesterol Precursor Profiles in N2a Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Durães, F.; Pinto, M.; Sousa, E. Old Drugs as New Treatments for Neurodegenerative Diseases. Pharmaceuticals 2018, 11, 44. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, C.; Moreira, P.I. Oxidative stress: A major player in cerebrovascular alterations associated to neurodegenerative events. Front. Physiol. 2018, 9, 806. [Google Scholar] [CrossRef] [PubMed]
- Franco-Iborra, S.; Vila, M.; Perier, C. Mitochondrial quality control in neurodegenerative diseases: Focus on Parkinson’s disease and Huntington’s disease. Front. Neurosci. 2018, 12, 342. [Google Scholar] [CrossRef] [PubMed]
- Elfawy, H.A.; Das, B. Crosstalk between mitochondrial dysfunction, oxidative stress, and age related neurodegenerative disease: Etiologies and therapeutic strategies. Life Sci. 2019, 218, 165–184. [Google Scholar] [CrossRef] [PubMed]
- Cummings, J.; Lee, G.; Ritter, A.; Zhong, K. Alzheimer’s disease drug development pipeline: 2018. Alzheimers Dement. (N. Y.) 2018, 4, 195–214. [Google Scholar] [CrossRef] [PubMed]
- Yeung, A.W.K.; Tzvetkov, N.T.; Atanasov, A.G. When Neuroscience Meets Pharmacology: A Neuropharmacology Literature Analysis. Front. Neurosci. 2018, 12, 852. [Google Scholar] [CrossRef] [PubMed]
- Pandareesh, M.D.; Mythri, R.B.; Srinivas Bharath, M.M. Bioavailability of dietary polyphenols: Factors contributing to their clinical application in CNS diseases. Neurochem. Int. 2015, 89, 198–208. [Google Scholar] [CrossRef]
- Kujawska, M.; Jodynis-Liebert, J. Polyphenols in Parkinson’s disease: A systematic review of in vivo studies. Nutrients 2018, 10, 642. [Google Scholar] [CrossRef]
- Namsi, A.; Nury, T.; Hamdouni, H.; Yammine, A.; Vejux, A.; Vervandier-Fasseur, D.; Latruffe, N.; Masmoudi-Kouki, O.; Lizard, G. Induction of neuronal differentiation of murine N2a cells by two polyphenols present in the Mediterranean diet mimicking neurotrophins activities: Resveratrol and apigenin. Diseases 2018, 6, 67. [Google Scholar] [CrossRef]
- Akagi, M.; Matsui, N.; Akae, H.; Hirashima, N.; Fukuishi, N.; Fukuyama, Y.; Akagi, R. Nonpeptide neurotrophic agents useful in the treatment of neurodegenerative diseases such as Alzheimer’s disease. J. Pharmacol. Sci. 2015, 127, 155–163. [Google Scholar] [CrossRef]
- Moosavi, F.; Hosseini, R.; Saso, L.; Firuzi, O. Modulation of neurotrophic signaling pathways by polyphenols. Drug Des. Dev. Ther. 2015, 10, 23–42. [Google Scholar]
- Murphy, K.E.; Park, J.J. Can Co-activation of Nrf2 and neurotrophic signaling pathway slow Alzheimer’s disease? Int. J. Mol. Sci. 2017, 18, 1168. [Google Scholar] [CrossRef] [PubMed]
- Sampaio, T.B.; Savall, A.S.; Gutierrez, M.E.Z.; Pinton, S. Neurotrophic factors in Alzheimer’s and Parkinson’s diseases: Implications for pathogenesis and therapy. Neural Regen. Res. 2017, 12, 549–557. [Google Scholar] [PubMed]
- Tonon, M.C.; Désy, L.; Nicolas, P.; Vaudry, H.; Pelletier, G. Immunocytochemical localization of the endogenous benzodiazepine ligand octadecaneuropeptide (ODN) in the rat brain. Neuropeptides 1990, 15, 17–24. [Google Scholar] [CrossRef]
- Alfonso, J.; Le Magueresse, C.; Zuccotti, A.; Khodosevich, K.; Monyer, H. Diazepam binding inhibitor promotes progenitor proliferation in the postnatal SVZ by reducing GABA signaling. Cell Stem Cell 2012, 10, 76–87. [Google Scholar] [CrossRef]
- Lacor, P.; Benavides, J.; Ferzaz, B. Enhanced expression of the peripheral benzodiazepine receptor (PBR) and its endogenous ligand octadecaneuropeptide (ODN) in the regenerating adult rat sciatic nerve. Neurosci. Lett. 1996, 220, 61–65. [Google Scholar] [CrossRef]
- Hamdi, Y.; Kaddour, H.; Vaudry, D.; Leprince, J.; Zarrouk, A.; Hammami, M.; Vaudry, H.; Tonon, M.C.; Amri, M.; Masmoudi-Kouki, O. Octadecaneuropeptide ODN prevents hydrogen, peroxide-induced oxidative damage of biomolecules in cultured rat astrocytes. Peptides 2015, 71, 56–65. [Google Scholar] [CrossRef]
- Ghouili, I.; Bahdoudi, S.; Morin, F.; Amri, F.; Hamdi, Y.; Coly, P.M.; Walet-Balieu, M.L.; Leprince, J.; Zekri, S.; Vaudry, H.; et al. Endogenous expression of ODN-related peptides in astrocytes contributes to cell protection against oxidative stress: Astrocyte-neuron crosstalk relevance for neuronal survival. Mol. Neurobiol. 2018, 55, 4596–4611. [Google Scholar] [CrossRef]
- Kaddour, H.; Hamdi, Y.; Vaudry, D.; Basille, M.; Desrues, L.; Leprince, J.; Castel, H.; Vaudry, H.; Tonon, M.C.; Amri, M.; et al. The octadecaneuropeptide ODN prevents 6-hydroxydopamine-induced apoptosis of cerebellar granule neurons through a PKC-MAPK-dependent pathway. J. Neurochem. 2013, 125, 620–633. [Google Scholar] [CrossRef]
- Kaddour, H.; Hamdi, Y.; Amri, F.; Bahdoudi, S.; Bouannee, I.; Leprince, J.; Zekri, S.; Vaudry, H.; Tonon, M.C.; Vaudry, D.; et al. Antioxidant and anti-apoptotic activity of octadecaneuropeptide against 6-OHDA toxicity in cultured at astrocytes. J. Mol. Neurosci. 2018, 69, 1–16. [Google Scholar] [CrossRef]
- Bahdoudi, S.; Ghouili, I.; Hmiden, M.; do Rego, J.L.; Lefranc, B.; Leprince, J.; Chuquet, J.; do Rego, J.C.; Marcher, A.B.; Mandrup, S.; et al. Neuroprotective effects of the gliopeptide ODN in an in vivo model of Parkinson’s disease. Cell. Mol. Life Sci. 2018, 75, 2075–2091. [Google Scholar] [CrossRef] [PubMed]
- Masmoudi-Kouki, O.; Hamdi, Y.; Ghouili, I.; Bahdoudi, S.; Kaddour, H.; Leprince, J.; Castel, H.; Vaudry, H.; Amri, M.; Vaudry, D.; et al. Neuroprotection with the Endozepine Octadecaneuropeptide, ODN. Curr. Pharm. Des. 2018, 24, 3918–3925. [Google Scholar] [CrossRef] [PubMed]
- Hamdi, Y.; Kaddour, H.; Vaudry, D.; Bahdoudi, S.; Douiri, S.; Leprince, J.; Castel, H.; Vaudry, H.; Tonon, M.C.; Amri, M.; et al. The octadecaneuropeptide ODN protects astrocytes against hydrogen peroxide-induced apoptosis via a PKA/MAPK-dependent mechanism. PLoS ONE 2012, 7, e42498. [Google Scholar] [CrossRef] [PubMed]
- Depreter, M.; Espeel, M.; Roels, F. Human peroxisomal disorders. Microsc. Res. Tech. 2003, 61, 203–223. [Google Scholar] [CrossRef] [PubMed]
- Lismont, C.; Nordgren, M.; Van Veldhoven, P.P.; Fransen, M. Redox interplay between mitochondria and peroxisomes. Front. Cell Dev. Biol. 2015, 3, 35. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Van Veldhoven, P.P.; Brees, C.; Rubio, N.; Nordgren, M.; Apanasets, O.; Kunze, M.; Baes, M.; Agostinis, P.; Fransen, M. Mitochondria are targets for peroxisome-derived oxidative stress in cultured mammalian cells. Free Radic Biol Med. 2013, 65, 882–894. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fransen, M.; Lismont, C.; Walton, P. The Peroxisome-mitochondria connection: How and why? Int. J. Mol. Sci. 2017, 18, 1126. [Google Scholar] [CrossRef]
- Berger, J.; Dorninger, F.; Forss-Petter, S.; Kunze, M. Peroxisomes in brain development and function. Biochim. Biophys. Acta 2016, 1863, 934–955. [Google Scholar] [CrossRef]
- Lee, A.; Hirabayashi, Y.; Kwon, S.K.; Lewis, T.L., Jr.; Polleux, F. Emerging roles of mitochondria in synaptic transmission and neurodegeneration. Curr. Opin. Physiol. 2018, 3, 82–93. [Google Scholar] [CrossRef]
- Baumgart, E.; Vanhorebeek, I.; Grabenbauer, M.; Borgers, M.; Declercq, P.E.; Fahimi, H.D.; Baes, M. Mitochondrial alterations caused by defective peroxisomal biogenesis in a mouse model for Zellweger syndrome (PEX5 knockout mouse). Am. J. Pathol. 2001, 159, 1477–1494. [Google Scholar] [CrossRef]
- Dirkx, R.; Vanhorebeek, I.; Martens, K.; Schad, A.; Grabenbauer, M.; Fahimi, D.; Declercq, P.; Van Veldhoven, P.P.; Baes, M. Absence of peroxisomes in mouse hepatocytes causes mitochondrial and ER abnormalities. Hepatology 2005, 41, 868–878. [Google Scholar] [CrossRef] [PubMed]
- Peeters, A.; Fraisl, P.; van den Berg, S.; Ver Loren van Themaat, E.; Van Kampen, A.; Rider, M.H.; Takemori, H.; van Dijk, K.W.; Van Veldhoven, P.P.; Carmeliet, P.; et al. Carbohydrate metabolism is perturbed in peroxisome-deficient hepatocytes due to mitochondrial dysfunction, AMP-activated protein kinase (AMPK) activation, and peroxisome proliferator-activated receptor γ coactivator 1α (PGC-1α) suppression. J. Biol. Chem. 2011, 286, 42162–42179. [Google Scholar] [CrossRef] [PubMed]
- Peeters, A.; Shinde, A.B.; Dirkx, R.; Smet, J.; De Bock, K.; Espeel, M.; Vanhorebeek, I.; Vanlander, A.; Van Coster, R.; Carmeliet, P.; et al. Mitochondria in peroxisome-deficient hepatocytes exhibit impaired respiration, depleted DNA, and PGC-1α independent proliferation. Biochim. Biophys. Acta 2015, 1853, 285–298. [Google Scholar] [CrossRef] [PubMed]
- López-Erauskin, J.; Galino, J.; Ruiz, M.; Cuezva, J.M.; Fabregat, I.; Cacabelos, D.; Boada, J.; Martínez, J.; Ferrer, I.; Pamplona, R.; et al. Impaired mitochondrial oxidative phosphorylation in the peroxisomal disease X-linked adrenoleukodystrophy. Hum. Mol. Genet. 2013, 22, 3296–3305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, P.; Wang, Q.; Yin, Y.; Yang, Z.; Li, W.; Liang, D.; Zhou, P. Association between peroxisome proliferator-activated receptor gamma gene polymorphisms and atherosclerotic diseases: A meta-analysis of case-control studies. J. Atheroscler. Thromb. 2015, 22, 912–925. [Google Scholar] [CrossRef] [PubMed]
- Cabezas-Opazo, F.A.; Vergara-Pulgar, K.; Pérez, M.J.; Jara, C.; Osorio-Fuentealba, C.; Quintanilla, R.A. Mitochondrial dysfunction contributes to the pathogenesis of Alzheimer’s disease. Oxid. Med. Cell. Longev. 2015, 2015, 509654. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, M.; Rodriguez-Sabate, C.; Morales, I.; Sanchez, A.; Sabate, M. Parkinson’s disease as a result of aging. Aging Cell 2015, 14, 293–308. [Google Scholar] [CrossRef]
- Ruffoli, R.; Bartalucci, A.; Frati, A.; Fornai, F. Ultrastructural studies of ALS mitochondria connect altered function and permeability with defects of mitophagy and mitochondriogenesis. Front. Cell. Neurosci. 2015, 9, 341. [Google Scholar] [CrossRef] [Green Version]
- Mamelak, M. Energy and the Alzheimer brain. Neurosci. Biobehav. Rev. 2017, 75, 297–313. [Google Scholar] [CrossRef]
- Campbell, G.; Mahad, D.J. Mitochondrial dysfunction and axon degeneration in progressive multiple sclerosis. FEBS Lett. 2018, 592, 1113–1121. [Google Scholar] [CrossRef]
- Lizard, G.; Rouaud, O.; Demarquoy, J.; Cherkaoui-Malki, M.; Iuliano, L. Potential roles of peroxisomes in Alzheimer’s disease and in dementia of the Alzheimer’s type. J. Alzheimers Dis. 2012, 29, 241–254. [Google Scholar] [CrossRef] [PubMed]
- Gray, E.; Rice, C.; Hares, K.; Redondo, J.; Kemp, K.; Williams, M.; Brown, A.; Scolding, N.; Wilkins, A. Reductions in neuronal peroxisomes in multiple sclerosis grey matter. Mult. Scler. 2014, 20, 651–659. [Google Scholar] [CrossRef] [PubMed]
- Gray, E.; Kemp, K.; Hares, K.; Redondo, J.; Rice, C.; Scolding, N.; Wilkins, A. Increased microglial catalase activity in multiple sclerosis grey matter. Brain Res. 2014, 1559, 55–64. [Google Scholar] [CrossRef]
- Senanayake, V.K.; Jin, W.; Mochizuki, A.; Chitou, B.; Goodenowe, D.B. Metabolic dysfunctions in multiple sclerosis: Implications as to causation, early detection, and treatment, a case control study. BMC Neurol. 2015, 15, 154. [Google Scholar] [CrossRef] [PubMed]
- Zarrouk, A.; Riedinger, J.M.; Ahmed, S.H.; Hammami, S.; Chaabane, W.; Debbabi, M.; Ben Ammou, S.; Rouaud, O.; Frih, M.; Lizard, G.; et al. Fatty acid profiles in demented patients: Identification of hexacosanoic acid (C26:0) as a blood lipid biomarker of dementia. J. Alzheimers Dis. 2015, 44, 1349–1359. [Google Scholar] [CrossRef] [PubMed]
- Zarrouk, A.; Debbabi, M.; Bezine, M.; Karym, E.M.; Badreddine, A.; Rouaud, O.; Moreau, T.; Cherkaoui-Malki, M.; El Ayeb, M.; Nasser, B.; et al. Lipid Biomarkers in Alzheimer’s Disease. Curr. Alzheimer Res. 2018, 15, 303–312. [Google Scholar] [CrossRef]
- Islinger, M.; Voelkl, A.; Fahimi, H.D.; Schrader, M. The peroxisome: An update on mysteries 2.0. Histochem. Cell Biol. 2018, 150, 443–471. [Google Scholar] [CrossRef]
- Schrader, M.; Costello, J.; Godinho, L.F.; Islinger, M. Peroxisome-mitochondria interplay and disease. J. Inherit. Metab. Dis. 2015, 38, 681–702. [Google Scholar] [CrossRef]
- Agostini, M.; Romeo, F.; Inoue, S.; Niklison-Chirou, M.V.; Elia, A.J.; Dinsdale, D.; Morone, N.; Knight, R.A.; Mak, T.W.; Melino, G. Metabolic reprogramming during neuronal differentiation. Cell Death Differ. 2016, 23, 1502–1514. [Google Scholar] [CrossRef] [Green Version]
- Costa, E.; Guidotti, A. Diazepam binding inhibitor (DBI): A peptide with multiple biological actions. Life Sci. 1991, 49, 325–344. [Google Scholar] [CrossRef]
- Vaudry, D.; Falluel-Morel, A.; Bourgault, S.; Basille, M.; Burel, D.; Wurtz, O.; Fournier, A.; Chow, B.K.; Hashimoto, H.; Galas, L.; et al. Pituitary adenylate cyclase activating polypeptide and its receptors: 20 years after the discovery. Pharmacol. Rev. 2009, 61, 283–357. [Google Scholar] [CrossRef] [PubMed]
- Hirabayashi, T.; Nakamachi, T.; Shioda, S. Discovery of PACAP and its receptors in the brain. J. Headache Pain 2018, 19, 28. [Google Scholar] [CrossRef] [PubMed]
- Vaudry, D.; Chen, Y.; Ravni, A.; Hamelink, C.; Elkahloun, A.G.; Eiden, L.E. Analysis of the PC12 cell transcriptome after differentiation with pituitary adenylate cyclase-activating polypeptide (PACAP). J. Neurochem. 2002, 83, 1272–1284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manecka, D.L.; Mahmood, S.F.; Grumolato, L.; Lihrmann, I.; Anouar, Y. Pituitary adenylate cyclase-activating polypeptide (PACAP) promotes both survival and neuritogenesis in PC12 cells through activation of nuclear factor κB (NF-κB) pathway: Involvement of extracellular signal-regulated kinase (ERK), calcium, and c-REL. J. Biol. Chem. 2013, 288, 14936–14948. [Google Scholar] [CrossRef] [PubMed]
- Chao, M.V.; Rajagopal, R.; Lee, F.S. Neurotrophin signalling in health and disease. Clin. Sci. (Lond.) 2006, 110, 167–173. [Google Scholar] [CrossRef] [PubMed]
- Leprince, J.; Oulyadi, H.; Vaudry, D.; Masmoudi, O.; Gandolfo, P.; Patte, C.; Costentin, J.; Fauchère, J.L.; Davoust, D.; Vaudry, H.; et al. Synthesis, conformational analysis and biological activity of cyclic analogs of the octadecaneuropeptide ODN. Design of a potent endozepine antagonist. Eur. J. Biochem. 2001, 268, 6045–6057. [Google Scholar] [CrossRef] [PubMed]
- Kharroubi, W.; Ahmed, S.H.; Nury, T.; Andreoletti, P.; Haouas, Z.; Zarrouk, A.; Sakly, R.; Hammami, M.; Lizard, G. Evidence of hormesis on human neuronal SK-N-BE cells treated with sodium arsenate: Impact at the mitochondrial level. Environ. Sci. Pollut. Res. Int. 2016, 23, 8441–8452. [Google Scholar] [CrossRef]
- Nury, T.; Sghaier, R.; Zarrouk, A.; Ménétrier, F.; Uzun, T.; Leoni, V.; Caccia, C.; Meddeb, W.; Namsi, A.; Sassi, K.; et al. Induction of peroxisomal changes in oligodendrocytes treated with 7-ketocholesterol: Attenuation by α-tocopherol. Biochimie 2018, 153, 181–202. [Google Scholar] [CrossRef]
- Schnell, C.; Hagos, Y.; Hülsmann, S. Active sulforhodamine 101 uptake into hippocampal astrocytes. PLoS ONE 2012, 7, e49398. [Google Scholar] [CrossRef]
- Moilanen, T.; Nikkari, T. The effect of storage on the fatty acid composition of human serum. Clin. Chim. Acta 1981, 114, 111–116. [Google Scholar] [CrossRef]
- Morrison, W.R.; Smith, L.M. Preparation of fatty acid methyl esters and dimethylacetals from lipids with boron fluoride–methanol. J. Lipid Res. 1964, 5, 600–608. [Google Scholar] [PubMed]
- Debbabi, M.; Nury, T.; Zarrouk, A.; Mekahli, N.; Bezine, M.; Sghaier, R.; Grégoire, S.; Martine, L.; Durand, P.; Camus, E.; et al. Protective effects of -tocopherol, -tocopherol and oleic acid, three compounds of olive oils, and no effect of trolox, on 7-ketocholesterol-induced mitochondrial and peroxisomal dysfunction in microglial BV-2 cells. Int. J. Mol. Sci. 2016, 17, 1973. [Google Scholar] [CrossRef] [PubMed]
- Badreddine, A.; Zarrouk, A.; Karym, E.M.; Debbabi, M.; Nury, T.; Meddeb, W.; Sghaier, R.; Bezine, M.; Vejux, A.; Martine, L.; et al. Argan Oil-Mediated Attenuation of Organelle Dysfunction, Oxidative Stress and Cell Death Induced by 7-Ketocholesterol in Murine Oligodendrocytes 158N. Int. J. Mol. Sci. 2017, 18, 2220. [Google Scholar] [CrossRef] [PubMed]
- Mitsche, M.A.; McDonald, J.G.; Hobbs, H.H.; Cohen, J.C. Flux analysis of cholesterol biosynthesis in vivo reveals multiple tissue and cell-type specific pathways. Elife 2015, 4, e07999. [Google Scholar] [CrossRef] [PubMed]
- Leoni, V.; Strittmatter, L.; Zorzi, G.; Zibordi, F.; Dusi, S.; Garavaglia, B.; Venco, P.; Caccia, C.; Souza, A.L.; Deik, A.; et al. Metabolic consequences of mitochondrial coenzyme A deficiency in patients with PANK2 mutations. Mol. Genet. Metab. 2011, 105, 463–471. [Google Scholar] [CrossRef] [PubMed]
- Leoni, V.; Nury, T.; Vejux, A.; Zarrouk, A.; Caccia, C.; Debbabi, M.; Fromont, A.; Sghaier, R.; Moreau, T.; Lizard, G. Mitochondrial dysfunctions in 7-ketocholesterol-treated 158N oligodendrocytes without or with α-tocopherol: Impacts on the cellular profil of tricarboxylic cycle-associated organic acids, long chain saturated and unsaturated fatty acids, oxysterols, cholesterol and cholesterol precursors. J. Steroid Biochem. Mol. Biol. 2017, 169, 96–110. [Google Scholar]
- Engelhard, H.H. Flow cytometric applications of Sulforhodamine 101 as a fluorescent stain for total cellular protein. Biotech. Histochem. 1997, 72, 1–9. [Google Scholar] [CrossRef]
- Costello, J.L.; Passmore, J.B.; Islinger, M.; Schrader, M. Multi-localized Proteins: The Peroxisome-Mitochondria Connection. Subcell. Biochem. 2018, 89, 383–415. [Google Scholar]
- Schrader, M.; Fahimi, H.D. Growth and division of peroxisomes. Int. Rev. Cytol. 2006, 255, 237–290. [Google Scholar]
- Schrader, M.; Fahimi, H.D. The peroxisome: Still a mysterious organelle. Histochem Cell Biol. 2008, 129, 421–440. [Google Scholar] [CrossRef]
- Trompier, D.; Vejux, A.; Zarrouk, A.; Gondcaille, C.; Geillon, F.; Nury, T.; Savary, S.; Lizard, G. Brain peroxisomes. Biochimie 2014, 98, 102–110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wanders, R.J.; Waterham, H.R. Biochemistry of mammalian peroxisomes revisited. Annu Rev. Biochem. 2006, 75, 295–332. [Google Scholar] [CrossRef] [PubMed]
- Pandak, W.M.; Ren, S.; Marques, D.; Hall, E.; Redford, K.; Mallonee, D.; Bohdan, P.; Heuman, D.; Gil, G.; Hylemon, P. Transport of cholesterol into mitochondria is rate-limiting for bile acid synthesis via the alternative pathway in primary rat hepatocytes. J. Biol. Chem. 2002, 277, 48158–48164. [Google Scholar] [CrossRef] [PubMed]
- Wanders, R.J. Peroxisomes, lipid metabolism, and peroxisomal disorders. Mol. Genet. Metab. 2004, 83, 16–27. [Google Scholar] [CrossRef] [PubMed]
- Wanders, R.J. Metabolic functions of peroxisomes in health and disease. Biochimie 2014, 98, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Knobloch, M. The Role of Lipid Metabolism for Neural Stem Cell Regulation. Brain Plast. 2017, 3, 61–71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katakura, M.; Hashimoto, M.; Okui, T.; Shahdat, H.M.; Matsuzaki, K.; Shido, O. Omega-3 polyunsaturated Fatty acids enhance neuronal differentiation in cultured rat neural stem cells. Stem Cells Int. 2013, 2013, 490476. [Google Scholar] [CrossRef]
- Cartocci, V.; Segatto, M.; Di Tunno, I.; Leone, S.; Pfrieger, F.W.; Pallottini, V. Modulation of the isoprenoid/cholesterol biosynthetic pathway during neuronal differentiation in vitro. J. Cell. Biochem. 2016, 117, 2036–2044. [Google Scholar] [CrossRef]
- Malagon, M.; Vaudry, H.; Van Strien, F.; Pelletier, G.; Gracia-Navarro, F.; Tonon, M.C. Ontogeny of diazepam-binding inhibitor-related peptides (endozepines) in the rat brain. Neuroscience 1993, 57, 777–786. [Google Scholar] [CrossRef]
- Ebadi, M.; Bashir, R.M.; Heidrick, M.L.; Hamada, F.M.; Refaey, H.E.; Hamed, A.; Helal, G.; Baxin, M.D.; Cerutis, D.R.; Lassi, N.K. Neurotrophins and their receptors in nerve injury and repair. Neurochem. Int. 1997, 30, 347–374. [Google Scholar] [CrossRef]
- Botia, B.; Basille, M.; Allais, A.; Raoult, E.; Falluel-Morel, A.; Galas, L.; Jolivel, V.; Wurtz, O.; Komuro, H.; Fournier, A.; et al. Neurotrophic effects of PACAP in the cerebellar cortex. Peptides 2007, 28, 1746–1752. [Google Scholar] [CrossRef] [PubMed]
- Masmoudi-Kouki, O.; Douiri, S.; Hamdi, Y.; Kaddour, H.; Bahdoudi, S.; Vaudry, D.; Basille, M.; Leprince, J.; Fournier, A.; Vaudry, H.; et al. Pituitary adenylate cyclase-activating polypeptide protects astroglial cells against oxidative stress-induced apoptosis. J. Neurochem. 2011, 117, 403–411. [Google Scholar] [CrossRef] [PubMed]
- Blondel, O.; Collin, C.; McCarran, W.J.; Zhu, S.; Zamostiano, R.; Gozes, I.; Brenneman, D.E.; McKay, R.D. A glia-derived signal regulating neuronal differentiation. J Neurosci. 2000, 20, 8012–8020. [Google Scholar] [CrossRef] [PubMed]
- Oz, S.; Ivashko-Pachima, Y.; Gozes, I. The ADNP derived peptide, NAP modulates the tubulin pool: Implication for neurotrophic and neuroprotective activities. PLoS ONE 2012, 7, e51458. [Google Scholar] [CrossRef] [PubMed]
- Greene, L.A.; McGuire, J.C. Induction of ornithine decarboxylase by nerve growth factor dissociated from effects on survival and neurite outgrowth. Nature 1978, 276, 191–194. [Google Scholar] [CrossRef] [PubMed]
- Noh, K.T.; Son, K.H.; Jung, I.D.; Kang, H.K.; Hwang, S.A.; Lee, W.S.; You, J.C.; Park, Y.M. Protein kinase C δ (PKCδ)-extracellular signal-regulated kinase 1/2 (ERK1/2) signaling cascade regulates glycogen synthase kinase-3 (GSK-3) inhibition-mediated interleukin-10 (IL-10) expression in lipopolysaccharide (LPS)-induced endotoxemia. J. Biol. Chem. 2012, 287, 14226–14233. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Dai, Y.B.; Sun, J.Y.; Xiang, Y.; Yang, J.; Dai, S.Y.; Zhang, X. Neuroglobin Attenuates Beta Amyloid-Induced Apoptosis Through Inhibiting Caspases Activity by Activating PI3K/Akt Signaling Pathway. J. Mol. Neurosci. 2016, 58, 28–38. [Google Scholar] [CrossRef] [PubMed]
- Algarni, A.S.; Hargreaves, A.J.; Dickenson, J.M. Activation of transglutaminase 2 by nerve growth factor in differentiating neuroblastoma cells: A role in cell survival and neurite outgrowth. Eur. J. Pharmacol. 2018, 820, 113–129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Almeida, A.S.; Vieira, H.L.A. Role of Cell Metabolism and Mitochondrial Function During Adult Neurogenesis. Neurochem. Res. 2017, 42, 1787–1794. [Google Scholar] [CrossRef] [PubMed]
- Campbell, G.R.; Worrall, J.T.; Mahad, D.J. The central role of mitochondria in axonal degeneration in multiple sclerosis. Mult. Scler. J. 2014, 20, 1806–1813. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sedel, F.; Bernard, D.; Mock, D.M.; Tourbah, A. Targeting demyelination and virtual hypoxia with high-dose biotin as a treatment for progressive multiple sclerosis. Neuropharmacology 2016, 110, 644–653. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Savary, S.; Trompier, D.; Andréoletti, P.; Le Borgne, F.; Demarquoy, J.; Lizard, G. Fatty acids—Induced lipotoxicity and inflammation. Curr. Drug Metab. 2012, 13, 1358–1370. [Google Scholar] [CrossRef] [PubMed]
- Thiemann, M.; Schrader, M.; Völkl, A.; Baumgart, E.; Fahimi, H.D. Interaction of peroxisomes with microtubules. In vitro studies using a novel peroxisome-microtubule binding assay. Eur. J. Biochem. 2000, 267, 6264–6275. [Google Scholar] [CrossRef] [PubMed]
- Farr, R.L.; Lismont, C.; Terlecky, S.R.; Fransen, M. Peroxisome biogenesis in mammalian cells: The impact of genes and environment. Biochim. Biophys. Acta 2016, 1863, 1049–1060. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are available from the authors. |







| Control (0% FBS) | ODN (10−14 M)–0% FBS | |
|---|---|---|
| Undifferentiated cells (without neurites) | 87.8 ± 3.5% | 35.2 ± 4.2% |
| Differentiated cells (neurites 5–10 µm length) | 4.6 ± 2.8% | 30.4 ± 5.3% |
| Differentiated cells (one or more neurites > 10 µm length without or with neurites 5–10 µm length) | 8.6 ± 4.1% | 34.4 ± 3.3% |
| Treatments | ||
|---|---|---|
| Control | ODN (10−14 M) | |
| Saturated fatty acids | ||
| C12:0 | 0.57 ± 0.34 | 0.67 ± 0.15 |
| C14:0 | 6.04 ± 3.40 | 8.22 ± 6.00 |
| C15:0 | 0.33 ± 0.19 | 0.40 ± 0.24 |
| C16:0 | 37.95 ± 20.47 | 47.50 ± 27.36 |
| C17:0 | 0.28 ± 0.14 | 0.33 ± 0.16 |
| C18:0 | 19.40 ± 10.27 | 25.23 ± 15.70 |
| C19:0 | 0.03 ± 0.02 | 0.04 ± 0.02 |
| C20:0 | 0.34 ± 0.18 | 0.47 ± 0.31 |
| C21:0 | 0.01 ± 0.01 | 0.01 ± 0.01 |
| C22:0 | 0.07 ± 0.05 | 0.10 ± 0.07 |
| C23:0 | 0.01 ± 0.01 | 0.01 ± 0.01 |
| C24:0 | 0.22 ± 0.13 | 0.28 ± 0.21 |
| C25:0 | 0.01 ± 0.01 | 0.02 ± 0.01 |
| C26:0 | 0.10 ± 0.06 | 0.12 ± 0.09 |
| Mono-unsaturated fatty acids | ||
| C14:1 | 0.02 ± 0.01 | 0.03 ± 0.01 |
| C16:1 n-7 | 0.94 ± 0.56 | 1.28 ± 0.95 |
| C16:1 n-9 | 0.79 ± 0.46 | 1.05 ± 0.70 |
| C18:1 n-7 | 10.75 ± 6.48 | 15.39 ± 12.95 |
| C18:1 n-9 | 54.64 ± 25.64 | 69.53 ± 37.16 |
| C20:1 n-7 | 0.71 ± 0.37 | 1.10 ± 0.90 |
| C20:1 n-9 | 3.00 ± 1.66 | 4.78 ± 4.09 |
| C22:1 n-7 | 0.06 ± 0.03 | 0.09 ± 0.07 |
| C22:1 n-9 | 0.45 ± 0.27 | 0.68 ± 0.58 |
| C24:1 n-9 | 0.26 ± 0.16 | 0.37 ± 0.29 |
| C26:1 | 0.11 ± 0.06 | 0.14 ± 0.10 |
| Poly-unsaturated fatty acids | ||
| C18:2 n-6 | 0.79 ± 0.47 | 1.04 ± 0.80 |
| C18:3 n-3 | 0.01 ± 0.01 | 0.02 ± 0.01 |
| C18:3 n-6 | 0.04 ± 0.02 | 0.05 ± 0.03 |
| C20:2 n-6 | 1.17 ± 0.53 | 1.79 ± 1.24 |
| C20:3 n-6 | 0.45 ± 0.24 | 0.60 ± 0.40 |
| C20:3 n-9 | 3.42 ± 1.73 | 5.02 ± 3.64 |
| C20:4 n-6 | 1.97 ± 1.29 | 2.40 ± 1.51 |
| C20:5 n-3 | 0.42 ± 0.27 | 0.58 ± 0.51 |
| C22:2 n-6 | 0.75 ± 0.37 | 1.08 ± 0.74 |
| C22:4 n-6 | 0.21 ± 0.20 | 0.26 ± 0.18 |
| C22:5 n-3 | 0.83 ± 0.56 | 0.98 ± 0.57 |
| C22:5 n-6 | 0.01 ± 0.01 | 0.02 ± 0.01 |
| C22:6 n-3 | 1.37 ± 0.90 | 1.68 ± 1.07 |
| C24:4 | 0.00 ± 0.01 | 0.01 ± 0.01 |
| C24:5 | 0.01 ± 0.01 | 0.01 ± 0.01 |
| C24:6 n-3 | 0.08 ± 0.05 | 0.10 ± 0.08 |
| Total fatty acids | 148.62 ± 10.55 | 192.48 ± 13.28 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Namsi, A.; Nury, T.; Khan, A.S.; Leprince, J.; Vaudry, D.; Caccia, C.; Leoni, V.; Atanasov, A.G.; Tonon, M.-C.; Masmoudi-Kouki, O.; et al. Octadecaneuropeptide (ODN) Induces N2a Cells Differentiation through a PKA/PLC/PKC/MEK/ERK-Dependent Pathway: Incidence on Peroxisome, Mitochondria, and Lipid Profiles. Molecules 2019, 24, 3310. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules24183310
Namsi A, Nury T, Khan AS, Leprince J, Vaudry D, Caccia C, Leoni V, Atanasov AG, Tonon M-C, Masmoudi-Kouki O, et al. Octadecaneuropeptide (ODN) Induces N2a Cells Differentiation through a PKA/PLC/PKC/MEK/ERK-Dependent Pathway: Incidence on Peroxisome, Mitochondria, and Lipid Profiles. Molecules. 2019; 24(18):3310. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules24183310
Chicago/Turabian StyleNamsi, Amira, Thomas Nury, Amira. S. Khan, Jérôme Leprince, David Vaudry, Claudio Caccia, Valerio Leoni, Atanas G. Atanasov, Marie-Christine Tonon, Olfa Masmoudi-Kouki, and et al. 2019. "Octadecaneuropeptide (ODN) Induces N2a Cells Differentiation through a PKA/PLC/PKC/MEK/ERK-Dependent Pathway: Incidence on Peroxisome, Mitochondria, and Lipid Profiles" Molecules 24, no. 18: 3310. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules24183310