Chemical Compositions of Propolis from China and the United States and their Antimicrobial Activities Against Penicillium notatum
Abstract
:1. Introduction
2. Results
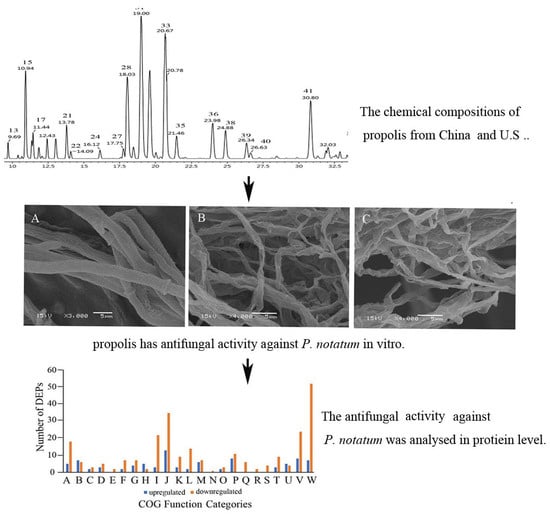
2.1. Chemical Composition
2.2. Antifungal Activities
2.3. Effects of EEP on Mycelial Growth
2.4. Effect of EEP on the Cell Membrane Permeability
2.5. Effect of EEP on the Activities of Succinate Dehydrogenase (SDH) and Malate Dehydrogenase (MDH)
2.6. iTRAQ Data Analysis and Differentially Expressed Proteins (DEPs) Identification
2.7. RT-PCR Analysis of the Differentially Expressed Proteins
3. Materials and Methods
3.1. Propolis Samples
3.2. Preparation of Experimental Samples
3.3. Total Polyphenol and Total Flavonoid Contents Determination
3.4. UPLC-ESI-MS Analysis
3.5. Antifungal Bioassay
3.6. Culture Conditions
3.7. Scanning Electron Microscope (SEM) Observation
3.8. Effects of EEP on mycelia Cell Membrane Permeability
3.9. Enzyme Activity Assays
3.10. iTRAQ Experiment
3.11. Quantitative Real-Time PCR
3.12. Statistical Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Athikomkulchai, S.; Awale, S.; Ruangrungsi, N.; Ruchirawat, S.; Kadota, S. Chemical constituents of Thai propolis. Fitoterapia 2013, 88, 96–100. [Google Scholar] [CrossRef] [PubMed]
- Popova, M.P.; Graikou, K.; Chinou, I.; Bankova, V.S. GC-MS Profiling of Diterpene Compounds in Mediterranean Propolis from Greece. J. Agric. Food Chem. 2010, 58, 3167–3176. [Google Scholar] [CrossRef] [PubMed]
- Funakoshi-Tago, M.; Okamoto, K.; Izumi, R.; Tago, K.; Yanagisawa, K.; Narukawa, Y.; Kiuchi, F.; Kasahara, T.; Tamura, H. Anti-inflammatory activity of flavonoids in Nepalese propolis is attributed to inhibition of the IL-33 signaling pathway. Int. Immunopharmacol. 2015, 25, 189–198. [Google Scholar] [CrossRef] [PubMed]
- Boonsai, P.; Phuwapraisirisan, P.; Chanchao, C. Antibacterial Activity of a Cardanol from Thai Apis mellifera Propolis. Int. J. Med. Sci. 2014, 11, 327–336. [Google Scholar] [CrossRef] [PubMed]
- Silva, M.S.S.; De Lima, S.G.; Oliveira, E.H.; Lopes, J.A.D.; Chaves, M.H.; Reis, F.A.M.; Citó, A.M.G.L. Anacardic acid derivatives from Brazilian propolis and their antibacterial activity. Eclética Química J. 2008, 33, 53–58. [Google Scholar] [CrossRef] [Green Version]
- Sanpa, S.; Popova, M.; Bankova, V.; Tunkasiri, T.; Eitssayeam, S.; Chantawannakul, P. Antibacterial Compounds from Propolis of Tetragonula laeviceps and Tetrigona melanoleuca (Hymenoptera: Apidae) from Thailand. PLoS ONE 2015, 10, e0126886. [Google Scholar] [CrossRef] [PubMed]
- Hamasaka, T.; Kumazawa, S.; Fujimoto, T.; Nakayama, T. Antioxidant Activity and Constituents of Propolis Collected in Various Areas of Japan. Food Sci. Technol. Res. 2004, 10, 86–92. [Google Scholar] [CrossRef] [Green Version]
- Hatano, A.; Nonaka, T.; Yoshino, M.; Ahn, M.-R.; Tazawa, S.; Araki, Y.; Kumazawa, S. Antioxidant Activity and Phenolic Constituents of Red Propolis from Shandong, China. Food Sci. Technol. Res. 2012, 18, 577–584. [Google Scholar] [CrossRef] [Green Version]
- Kalogeropoulos, N.; Konteles, S.J.; Troullidou, E.; Mourtzinos, I.; Karathanos, V.T. Chemical composition, antioxidant activity and antimicrobial properties of propolis extracts from Greece and Cyprus. Food Chem. 2009, 116, 452–461. [Google Scholar] [CrossRef]
- Laskar, R.A.; Sk, I.; Roy, N.; Begum, N.A. Antioxidant activity of Indian propolis and its chemical constituents. Food Chem. 2010, 122, 233–237. [Google Scholar] [CrossRef]
- Bonvehí, J.S.; Gutiérrez, A.L. The antimicrobial effects of propolis collected in different regions in the basque country (northern spain). World J. Microbiol. & Biotechnol. 2012, 28, 1351–1358. [Google Scholar]
- He, M.; Wu, T.; Pan, S.; Xu, X. Antimicrobial mechanism of flavonoids against Escherichia coli ATCC 25922 by model membrane study. Appl. Surf. Sci. 2014, 305, 515–521. [Google Scholar] [CrossRef]
- da Cunha, M.G.; Franchin, M.; de Carvalho Galvão, L.C.; de Ruiz, A.L.; de Carvalho, J.E.; Ikegaki, M.; de Alencar, S.M.; Koo, H.; Rosalen, P.L. Antimicrobial and antiproliferative activities of stingless bee Melipona scutellaris geopropolis. BMC Complement. Altern. Med. 2013, 13, 23. [Google Scholar] [CrossRef] [PubMed]
- Campos, J.F.; Dos Santos, U.P.; Macorini, L.F.B.; De Melo, A.M.M.F.; Balestieri, J.B.P.; Paredes-Gamero, E.J.; Cardoso, C.A.L.; Souza, K.D.P.; Dos Santos, E.L. Antimicrobial, antioxidant and cytotoxic activities of propolis from Melipona orbignyi (Hymenoptera, Apidae). Food Chem. Toxicol. 2014, 65, 374–380. [Google Scholar] [CrossRef] [PubMed]
- Peng, L.; Yang, S.; Cheng, Y.J.; Chen, F.; Pan, S.; Fan, G. Antifungal activity and action mode of pinocembrin from propolis against Penicillium italicum. Food Sci. Biotechnol. 2012, 21, 1533–1539. [Google Scholar] [CrossRef]
- De Castro, P.A.; Bom, V.L.P.; Brown, N.A.; De Almeida, R.S.C.; Ramalho, L.N.Z.; Savoldi, M.; Goldman, M.H.S.; Berretta, A.A.; Goldman, G.H. Identification of the cell targets important for propolis-induced cell death in Candida albicans. Fungal Genet. Boil. 2013, 60, 74–86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gatto, M.A.; Ippolito, A.; Linsalata, V.; Cascarano, N.A.; Nigro, F.; Vanadia, S.; Venere, D.D. Activity of extracts from wild edible herbs against postharvest fungal diseases of fruit and vegetables. Postharvest Biol. Technol. 2012, 61, 72–82. [Google Scholar] [CrossRef]
- Hou, Y.; Zheng, Z.; Xu, S.; Chen, C.; Zhou, M. Proteomic analysis of Fusarium graminearum treated by the fungicide JS399-19. Pestic. Biochem. Physiol. 2013, 107, 86–92. [Google Scholar] [CrossRef]
- Cagas, S.E.; Jain, M.R.; Li, H.; Perlin, D.S. Profiling the Aspergillus fumigatus proteome in response to caspofungin. Antimicrob. Agents & Chemother. 2011, 55, 146–154. [Google Scholar]
- Cicco, N.; Lanorte, M.; Paraggio, M.; Viggiano, M.; Lattanzio, V. A reproducible, rapid and inexpensive Folin–Ciocalteu micro-method in determining phenolics of plant methanol extracts. Microchem. J. 2013, 91, 107–110. [Google Scholar] [CrossRef]
- Falcao, S.I.; Vale, N.; Gomes, P.; Domingues, M.R.; Freire, C.; Cardoso, S.M.; Vilas-Boas, M. Phenolic Profiling of Portuguese Propolis by LC-MS Spectrometry: Uncommon Propolis Rich in Flavonoid Glycosides. Phytochemical Analysis. 2013, 24, 309–318. [Google Scholar] [CrossRef] [PubMed]
- Gardana, C.; Scaglianti, M.; Pietta, P.; Simonetti, P. Analysis of the polyphenolic fraction of propolis from different sources by liquid chromatography-tandem mass spectrometry. J. Pharm. Biomed. Anal. 2007, 45, 390–399. [Google Scholar] [CrossRef] [PubMed]
- Midorikawa, K.; Banskota, A.H.; Tezuka, Y.; Nagaoka, T.; Matsushige, K.; Message, D.; Huertas, A.A.G.; Kadota, S. Liquid chromatography-mass spectrometry analysis of propolis. Phytochem. Anal. 2001, 12, 366–373. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, K.; Komura, S.; Isaji, N.; Ohishi, N.; Yagi, K. Isolation of antioxidative compounds from Brazilian propolis: 3,4-dihydroxy-5-prenylcinnamic acid, a novel potent antioxidant. Chem. Pharm. Bull. 1999, 47, 1521–1524. [Google Scholar] [CrossRef]
- Miguel, M.G.; Nunes, S.; Dandlen, S.A.; Cavaco, A.M.; Antunes, M.D. Phenols, flavonoids and antioxidant activity of aqueous and methanolic extracts of propolis (Apis mellifera L.) from Algarve, South Portugal. Food Sci. Technol. 2014, 34, 16–23. [Google Scholar] [CrossRef] [Green Version]
- de Souza, E.L.; de Barros, J.C.; de Oliveira, C.E.; da Conceição, M.L. Influence of Origanum vulgare L. essential oil on enterotoxin production, membrane permeability and surface characteristics of Staphylococcus aureus. Int. J. Food Microbiol. 2010, 137, 308–311. [Google Scholar] [CrossRef] [PubMed]
- Bonvehi, J.S.; Gutiérrez, A.L. Antioxidant Activity and Total Phenolics of Propolis from the Basque Country (Northeastern Spain). J. Am. Oil Chem. Soc. 2011, 88, 1387–1395. [Google Scholar] [CrossRef]
- Uzel, A.; Sorkun, K.; Onçağ, O.; Cogŭlu, D.; Gençay, O.; Sali˙h, B. Chemical compositions and antimicrobial activities of four different Anatolian propolis samples. Microbiol. Res. 2005, 160, 189–195. [Google Scholar] [CrossRef]
- Mercan, N.; Kivrak, I.; Duru, M.E.; Katircioglu, H.; Gulcan, S.; Malci, S.; Acar, G.; Salih, B.; Malcı, S. Chemical composition effects onto antimicrobial and antioxidant activities of propolis collected from different regions of Turkey. Ann. Microbiol. 2006, 56, 373–378. [Google Scholar] [CrossRef]
- Campana, R.; Patrone, V.; Franzini, I.T.M.; Diamantini, G.; Vittoria, E.; Baffone, W. Antimicrobial Activity of Two Propolis Samples Against Human Campylobacter jejuni. J. Med. Food 2009, 12, 1050–1056. [Google Scholar] [CrossRef]
- Yang, S.; Peng, L.; Su, X.; Chen, F.; Cheng, Y.; Fan, G.; Pan, S. Bioassay-guided isolation and identification of antifungal components from propolis against Penicillium italicum. Food Chem. 2011, 127, 210–215. [Google Scholar] [CrossRef]
- Salas, A.L.; Alberto, M.R.; Zampini, I.C.; Cuello, A.S.; Maldonado, L.; Ríos, J.L.; Schmeda-Hirschmann, G.; Isla, M.I. Biological activities of polyphenols-enriched propolis from Argentina arid regions. Phytomedicine 2016, 23, 27–31. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Cao, X.; Ping, S.; Wang, K.; Shi, J.; Zhang, C.; Zheng, H.; Hu, F. Comparisons of Ethanol Extracts of Chinese Propolis (Poplar Type) and Poplar Gums Based on the Antioxidant Activities and Molecular Mechanism. Evid. -Based Complementary Altern. Med. 2015, 2015, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, S.; Zhang, C.P.; Wang, K.; Li, G.Q.; Hu, F.L. Recent Advances in the Chemical Composition of Propolis. Molecules 2014, 19, 19610–19632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Popova, M.; Dimitrova, R.; Al-Lawati, H.; Tsvetkova, I.; Najdenski, H.; Bankova, V. Omani propolis: Chemical profiling, antibacterial activity and new propolis plant sources. Chem. Cent. J. 2013, 7, 158. [Google Scholar] [CrossRef] [PubMed]
- Abreu, A.C.; Mcbain, A.J.; Simões, M. Plants as sources of new antimicrobials and resistance-modifying agents. Nat. Prod. Rep. 2012, 29, 1007–1021. [Google Scholar] [CrossRef] [PubMed]
- Haghdoost, N.; Salehi, T.; Khosravi, A.; Sharifzadeh, A. Antifungal activity and influence of propolis against germ tube formation as a critical virulence attribute by clinical isolates of Candida albicans. J. De Mycol. Médicale 2016, 26, 298–305. [Google Scholar] [CrossRef] [PubMed]
- Yao, X.; Zhu, X.; Pan, S.; Fang, Y.; Jiang, F.; Phillips, G.O.; Xu, X. Antimicrobial activity of nobiletin and tangeretin against Pseudomonas. Food Chem. 2012, 132, 1883–1890. [Google Scholar] [CrossRef]
- Franklin, D.; Zhang, Y. Extra-Ribosome Functions of Ribosomal Proteins. Encycl. Cell Biol. 2016, 281–287. [Google Scholar]
- Zaher, H.S.; Green, R. Quality control by the ribosome following peptide bond formation. Nature. 2008, 457, 161–166. [Google Scholar] [CrossRef] [Green Version]
- Warner, J.R.; McIntosh, K.B. How common are extraribosomal functions of ribosomal proteins? Mol. Cell 2009, 34, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Lindström, M.S.; Zhang, Y. Ribosomal protein S9 is a novel B23/NPM-binding protein required for normal cell proliferation. J. Boil. Chem. 2008, 283, 15568–15576. [Google Scholar] [CrossRef] [PubMed]
- Hartman, T.; Weinrick, B.; Vilchèze, C.; Berney, M.; Tufariello, J.; Cook, G.M.; Jacobs, W.R. Succinate Dehydrogenase is the Regulator of Respiration in Mycobacterium tuberculosis. PLoS Pathog. 2014, 10, e1004510. [Google Scholar] [CrossRef] [PubMed]
- Pecsi, I.; Hards, K.; Ekanayaka, N.; Berney, M.; Hartman, T.; Jacobs, W.R., Jr.; Cook, G.M. Essentiality of succinate dehydrogenase in mycobacterium smegmatis and its role in the generation of the membrane potential under hypoxia. Mbio 2014, 5, e01093-14. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Liu, L.; Li, D.; Xia, H.; Su, X.; Peng, L.; Pan, S. Use of active extracts of poplar buds against Penicillium italicum and possible modes of action. Food Chem. 2016, 196, 610–618. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Tong, Y.; Tian, M.; Sun, X.; Wang, S.; Ding, C.; Yu, S. Characterization of the immunogenicity and pathogenicity of malate dehydrogenase in Brucella abortus. World J. Microbiol. Biotechnol. 2014, 30, 2063–2070. [Google Scholar] [CrossRef] [PubMed]
- Saradhi, P.P.; Alia; Vani, B. Inhibition of mitochondrial electron transport is the prime cause behind proline accumulation during mineral deficiency in Oryza sativa. Plant Soil 1993, 155, 465–468. [Google Scholar] [CrossRef]
- Puntel, R.L.; Roos, D.H.; Seeger, R.L.; Rocha, J.B.; Da Rocha, J.B.T. Mitochondrial electron transfer chain complexes inhibition by different organochalcogens. Toxicol. Vitr. 2013, 27, 59–70. [Google Scholar] [CrossRef]
- Le, S.B.; Hailer, M.K.; Buhrow, S.; Wang, Q.; Flatten, K.; Pediaditakis, P.; Bible, K.C.; Lewis, L.D.; Sausville, E.A.; Pang, Y.-P.; et al. Inhibition of Mitochondrial Respiration as a Source of Adaphostin-induced Reactive Oxygen Species and Cytotoxicity. J. Boil. Chem. 2007, 282, 8860–8872. [Google Scholar] [CrossRef] [Green Version]
- Obermeyer, G.; Fragner, L.; Lang, V.; Weckwerth, W. Dynamic adaption of metabolic pathways during germination and growth of lily pollen tubes after inhibition of the electron transport chain. Plant. Physiol. 2013, 162, 1822–1833. [Google Scholar] [CrossRef]
- Martins, V.D.P.; Dinamarco, T.M.; Curti, C.; Uyemura, S.A. Classical and alternative components of the mitochondrial respiratory chain in pathogenic fungi as potential therapeutic targets. J. Bioenerg. Biomembr. 2011, 43, 81–88. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.H.; Kim, S.H.; Kim, S.Z.; Park, W.H. Antimycin A as a mitochondrial electron transport inhibitor prevents the growth of human lung cancer A549 cells. Oncol. Rep. 2008, 20, 689–693. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Jin, M.; Cai, Y.; Xia, H.; Long, K.; Liu, J.; Yu, Q.; Yuan, J. Mitochondrial electron transport chain complex III is required for antimycin A to inhibit autophagy. Chem. Boil. 2011, 18, 1474–1481. [Google Scholar] [CrossRef] [PubMed]
- Taubitz, A.; Bauer, B.; Heesemann, J.; Ebel, F. Role of Respiration in the Germination Process of the Pathogenic Mold Aspergillus fumigatus. Curr. Microbiol. 2007, 54, 354–360. [Google Scholar] [CrossRef] [PubMed]
- Jaña, F.; Faini, F.; LaPier, M.; Pavani, M.; Kemmerling, U.; Morello, A.; Maya, J.D.; Jara, J.; Parra, E.; Ferreira, J. Tumor cell death induced by the inhibition of mitochondrial electron transport: The effect of 3-hydroxybakuchiol. Toxicol. Appl. Pharmacol. 2013, 272, 356–364. [Google Scholar] [CrossRef] [PubMed]
- Jing, L.; Kumari, S.; Mendelev, N.; Li, P.A. Coenzyme Q10 Ameliorates Ultraviolet B Irradiation Induced Cell Death Through Inhibition of Mitochondrial Intrinsic Cell Death Pathway. Int. J. Mol. Sci. 2011, 12, 8302–8315. [Google Scholar] [CrossRef]
- Heyland, J.; Fu, J.; Blank, L.M. Correlation between TCA cycle flux and glucose uptake rate during respiro-fermentative growth of Saccharomyces cerevisiae. Microbiol. 2009, 155, 3827–3837. [Google Scholar] [CrossRef] [Green Version]
- Blank, L.M.; Sauer, U. TCA cycle activity in Saccharomyces cerevisiae is a function of the environmentally determined specific growth and glucose uptake rates. Microbiol. 2004, 150, 1085–1093. [Google Scholar] [CrossRef] [Green Version]
- Patel, M.S.; Korotchkina, L.G. The biochemistry of the pyruvate dehydrogenase complex. Biochem. Mol. Biol. Educ. 2010, 31, 5–15. [Google Scholar]
- Gamarra, S.; Rocha, E.M.; Zhang, Y.Q.; Park, S.; Rao, R.; Perlin, D.S. Mechanism of the synergistic effect of amiodarone and fluconazole in Candida alhicans. Antimicrob. Agents Chemot. 2010, 54, 1753–1761. [Google Scholar] [CrossRef]
- Ghannoum, M.; Isham, N.; Verma, A.; Plaum, S.; Fleischer, A.; Hardas, B. In Vitro Antifungal Activity of Naftifine Hydrochloride against Dermatophytes. Antimicrob. Agents Chemother. 2013, 57, 4369–4372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nowosielski, M.; Hoffmann, M.; Wyrwicz, L.S.; Stepniak, P.; Plewczynski, D.M.; Lazniewski, M.; Ginalski, K.; Rychlewski, L. Detailed Mechanism of Squalene Epoxidase Inhibition by Terbinafine. J. Chem. Inf. Model. 2011, 51, 455–462. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Jiang, J.; Yin, Y.; Ma, Z. Involvement of FgERG4 in ergosterol biosynthesis, vegetative differentiation and virulence in Fusarium graminearum. Mol. Plant Pathol. 2013, 14, 71–83. [Google Scholar] [CrossRef] [PubMed]
- Long, N.; Xu, X.; Zeng, Q.; Sang, H.; Lu, L. Erg4A and Erg4B Are Required for Conidiation and Azole Resistance via Regulation of Ergosterol Biosynthesis in Aspergillus fumigatus. Appl. Environ. Microbiol. 2016, 83, e02924-16. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, S.; Esquivel, B.D.; White, T.C. Overexpression or Deletion of Ergosterol Biosynthesis Genes Alters Doubling Time, Response to Stress Agents, and Drug Susceptibility in Saccharomyces cerevisiae. mBio 2018, 9, e01291-18. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds 2, 10, 11, 28 are available from the authors. |






| Samples | Total Phenol (mg·g−1) | Total Flavonoids (mg·g−1) |
|---|---|---|
| EEP-A | 370.19 ± 4.12b | 197.93 ± 1.24c |
| EEP-C | 440.18 ± 5.11a | 305.60 ± 1.69b |
| No | tR (min) | λmax (nm) | Selected Ion | Formula | Measured Mass | Calculated Mass | Mass Error (MD) | MS/MS Fragmentation | Compound Name | Relative Area(%) |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2.66 | 238, 281 | [M − H] | C7H6O3 | 137.0245 | 137.0239 | 0.6 | 137.0245 | p-Hydroxybenzoic acid (b) | 0.02% (C) |
| 2 | 3.29 | 243, 324 | [M − H] | C9H8O4 | 179.0351 | 179.0344 | 0.7 | 179.0344, 161.0243, 135.0448 | Caffeic acid (b, c) | 0.06% (A), 0.45% (C) |
| 3 | 4.61 | 237, 310 | [M − H] | C9H8O3 | 163.0400 | 163.0395 | 0.5 | 163.0400, 119.0502, 93.0368 | p-Coumaric acid (b, c) | 1.31% (A), 0.15% (C) |
| 4 | 5.20 | 243,323 | [M − H] | C10H10O4 | 193.0505 | 193.0505 | 0 | 193.0505,133.0291 | Isoferulic acid (b, c) | 0.04% (C) |
| 5 | 7.71 | 265, 320 | [M − H] | C21H20O10 | 431.1013 | 431.0978 | 3.5 | 431.1013, 268.0394 | Apigenin-7-O-β-D-glucopyranoside (b) | 0.27% (A), 0.35% (C) |
| 6 | 8.29 | 250, 365 | [M − H] | C16H11O6 | 299.0581 | 299.0556 | 2.5 | 299.0581, 227.0363, 129.0349 | Kaempferol-methyl ether (b) | 0.13% (A), 0.09% (C) |
| 7 | 8.51 | 292 | [M − H] | C16H14O5 | 285.0783 | 285.0763 | 2.0 | 285.0783, 267.0668, 252.0437, 239.0722, 138.0326 | Pinobanksin-methyl ether (b) | 0.24% (C) |
| 8 | 8.63 | 250, 365 | [M − H] | C17H14O7 | 329.0683 | 329.0661 | 2.4 | 329.0683, 314.0441, 299.0209, 271.0246, 135.0458 | Quercetin-dimethyl ether (b) | 0.03% (A), 0.05% (C) |
| 9 | 8.92 | 292 | [M − H] | C16H14O5 | 285.0779 | 285.0763 | 1.6 | 269.04620, 139.0406, 124.0170 | Pinobanksin-5-methyl ether (b, c) | 0.13% (A), 0.15% (C) |
| 10 | 8.92 | 251, 349 | [M − H] | C15H10O6 | 285.0418 | 285.0399 | 1.9 | 269.0462, 151.0039 | Luteolin (a, b) | 0.13% (A), 0.15% (C) |
| 11 | 9.02 | 253, 372 | [M − H] | C15H10O7 | 301.0365 | 301.0348 | 1.7 | 243.02886, 151.00304 | Quercetin (a, b) | 0.15% (A), 0.17% (C) |
| 12 | 9.20 | 287 | [M − H] | C16H14O5 | 285.0778 | 285.0763 | 1.5 | 285.0778, 267.0667, 252.0432, 239.0716, 138.0325 | Pinobanksin-methyl ether isomer (b) | 5.36% (A), 4.24% (C) |
| 13 | 9.74 | 255, 352 | [M − H] | C16H11O7 | 315.0514 | 315.0505 | 0.9 | 315.0514, 300.0277 | Quercetin-3-methyl ether (b, c) | 0.61% (A), 1.00% (C) |
| 14 | 10.44 | 265, 312 | [M − H] | C16H12O4 | 267.0663 | 267.0657 | 0.6 | 252.0424, 224.0476 | Chrysin-5-methyl ether (b, c) | 0.74% (A), 0.21% (C) |
| 15 | 10.94 | 292 | [M − H] | C15H12O5 | 271.0613 | 271.0606 | 0.7 | 271.0613, 253.0504 | Pinobanksin (b, c) | 5.68% (A), 6.26% (C) |
| 16 | 11.40 | 265, 365 | [M − H] | C15H10O6 | 285.0400 | 285.0399 | 0.1 | 285.0400, 227.03402 | Kaempferol (a, b) | 0.74% (A), 0.21% (C) |
| 17 | 11.44 | 285 | [M − H] | C16H14O4 | 269.0813 | 269.0814 | −0.1 | 269.0813, 254.0571, 227.0701, 165.0186 | Pinocembrin-5-methyl ether (b, c) | 1.22% (A), 0.26% (C) |
| 18 | 11.90 | 251, 366 | [M − H] | C16H11O7 | 315.0504 | 315.0505 | −0.1 | 315.05045, 300.02677, 151.00311 | Isorhamnetin (b, c) | 0.60% (A), 0.53% (C) |
| 19 | 12.43 | 266, 349 | [M − H] | C16H11O6 | 299.0555 | 299.0556 | −0.1 | 299.0555, 284.03186 | Kaempferol-3-methyl ether (b, c) | 1.08% (A), 0.99% (C) |
| 20 | 13.06 | 251, 351 | [M − H] | C17H14O7 | 329.0664 | 329.0661 | 0.3 | 329.0664, 314.0426, 299.0193, 271.0246, 133.0291 | Quercetin-dimethyl ether isomer (b, c) | 1.30% (A), 1.29% (C); |
| 21 | 13.85 | 259, 350 | [M − H] | C16H12O5 | 283.0618 | 283.0606 | 1.2 | 283.0618, 268.0379, 239.0352 | Galangin-5-methyl ether (b, c) | 2.10% (A), 2.56% (C) |
| 22 | 14.13 | 292 | [M − H] | C18H16O6 | 327.0880 | 327.0869 | 1.1 | 285.0768, 252.0429 | Pinobanksin-5-methyl ether-3-O-acetate (b, c) | 0.41% (A), 0.26% (C) |
| 23 | 14.94 | 251, 365 | [M − H] | C16H11O6 | 299.0557 | 299.0556 | 0.1 | 299.0557, 284.0322, 151.0027 | Kaempferol-7-methyl ether (b) | 0.11% (C) |
| 24 | 16.12 | 251, 366 | [M − H] | C17H14O7 | 329.0665 | 329.0661 | 0.4 | 329.0660, 314.0411, 299.0186, 271.0246, 161.0235 | Quercetin-dimethyl ether isomer (b) | 0.57% (A) |
| 25 | 16.55 | 251,351 | [M − H] | C17H14O7 | 329.0664 | 329.0661 | 0.3 | 329.0664, 314.0426, 299.0193, 271.0246, 133.0292 | Quercetin-dimethyl ether isomer (b) | 0.79% (C) |
| 26 | 17.53 | 324 | [M − H] | C14H16O4 | 247.0971 | 247.0970 | 0.1 | 247.0971, 179.0341, 161.0236, 135.0443 | Caffeic acid isoprenyl ester (b, c) | 2.75% (C), |
| 27 | 17.75 | 251,350 | [M − H] | C18H16O7 | 343.0818 | 343.0818 | 0 | 343.0818, 328.0579, 313.0345, 298.0113 | Quercetin-trimethyl ether (b) | 0.69% (A) |
| 28 | 18.03 | 267, 313 | [M − H] | C15H10O4 | 253.0511 | 253.0501 | 1.0 | 145.0294, 107.0138 | Chrysin (a, b) | 8.33% (A), 9.07% (C) |
| 29 | 18.25 | 324 | [M − H] | C14H16O4 | 247.0977 | 247.0970 | 0.7 | 179.0346, 161.0241, 135.0449 | Caffeic acid isoprenyl ester isomer (b, c) | 1.48% (C) |
| 30 | 18.52 | 324 | [M − H] | C16H14O4 | 269.0821 | 269.0814 | 0.7 | 269.0821, 179.9343m 133.0294 | Caffeic acid benzyl ester (b, c) | 0.88% (A), 1.91% (C) |
| 31 | 19.00 | 286 | [M − H] | C15H12O4 | 255.0662 | 255.0657 | 0.5 | 213.0552, 151.0031, 107.0134 | Pinocembrin (b, c) | 16.14% (A), 9.66% (C) |
| 32 | 19.65 | 265, 361 | [M − H] | C15H10O5 | 269.0456 | 269.0450 | 0.6 | 211.03914, 145.0288, 117.0340 | Galangin (a, b) | 9.74% (A), 9.90% (C) |
| 33 | 20.67 | 292 | [M − H] | C17H14O6 | 313.0745 | 313.0712 | 3.3 | 253.0505, 119.0498 | Pinobanksin-3-O-acetate (b, c) | 14.52% (A), 11.40% (C) |
| 34 | 20.88 | 324 | [M − H] | C17H16O4 | 283.0975 | 283.0970 | 0.5 | 179.0347, 161.0240, 135.0447 | Phenethyl caffeate (b,c) | 2.96% (C) |
| 35 | 21.46 | 265, 323 | [M − H] | C16H12O5 | 283.0611 | 283.0606 | 0.5 | 283.0611, 268.03733, 239.0346 | Acacetin (b, c) | 2.00% (A), 2.77% (C) |
| 36 | 23.98 | 310 | [M − H] | C16H14O3 | 253.0866 | 253.0865 | 0.1 | 145.0287, 117.0339 | p-Coumaric acid benzyl ester (b,d) | 3.46% (A) |
| 37 | 24.08 | 292 | [M − H] | C22H22O8 | 461.1248 | 461.1236 | 1.2 | 401.1012, 253.0874 | Pinobanksin-3-O-acetate -5-O-phydroxyphenylpropionate (b,c) | 0.48% (C) |
| 38 | 24.88 | 324 | [M − H] | C18H16O4 | 295.0984 | 295.0970 | 1.4 | 178.0270, 133.0296 | Caffeic acid cinnamyl ester (b, c) | 2.63% (A), 5.00% (C) |
| 39 | 26.42 | 292 | [M − H] | C18H16O6 | 327.0874 | 327.0869 | 0.5 | 271.0604, 253.0504 | Pinobanksin-3-O-propionate (b, c) | 1.41% (A), 5.50% (C) |
| 40 | 26.34 | 310 | [M − H] | C18H16O3 | 267.1021 | 267.1021 | 0 | 163.0390,145.0288, 119.0495 | p-Coumaric acid benylethyl ester (b) | 0.51% (A) |
| 41 | 30.80 | 310 | [M − H] | C18H16O3 | 279.1027 | 279.1021 | 0.6 | 235.1120, 134.0355, 163.0028 | p-Coumaric cinnamyl ester (b,d) | 6.25% (A) |
| 42 | 32.13 | 292 | [M − H] | C19H18O6 | 341.1036 | 341.1025 | 1.1 | 271.0610,253.0509 | Pinobanksin-3-O-butyrate (b,c) | 3.23% (C) |
| 43 | 32.65 | 292 | [M − H] | C20H18O6 | 353.1039 | 353.1025 | 1.4 | 297.1532, 253.0511 | Pinobanksin-3-O--pentenoate (b,c) | 0.72% (A) |
| 44 | 33.38 | 292 | [M − H] | C27H24O8 | 475.1397 | 475.1393 | 0.4 | 415.1174, 264.0500, 134.0366 | Pinobanksin-3-O-acetate-5-O-phydroxyphenylpropionate (b,c) | 0.18% (A) |
| 45 | 33.89 | 292 | [M − H] | C22H16O6 | 375.0894 | 375.0869 | 2.5 | 271.0603,253.0504 | Pinobanksin-3-O-benzoate (b) | 1.53% (A) |
| 46 | 35.98 | 292 | [M − H] | C20H20O6 | 355.1207 | 355.1182 | 2.5 | 253.0520, 271.0619 | Pinobanksin-3-O-pentanoate or 2-methylbutyrateb (b, c) | 0.13% (A), 2.85% (C) |
| 47 | 36.68 | 292 | [M − H] | C21H20O6 | 367.1205 | 367.1182 | 2.3 | 271.0624,253.0512 | Pinobanksin-O-hexenoate b,c | 0.56% (C) |
| 48 | 37.43 | 292 | [M − H] | C24H20O6 | 403.1200 | 403.1182 | 1.8 | 297.1132, 271.0614, 253.0513 | Pinobanksin-3-O-phenylpropionate (b, c) | 0.20% (A), 0.19% (C) |
| 49 | 38.69 | 292 | [M − H] | C21H22O6 | 369.1352 | 369.1338 | 1.4 | 271.0612, 253.0511 | Pinobanksin-3-O-hexanoate (b, c) | 1.08% (A), 0.84% (C) |
| Concentration (mg·mL−1) | MIC (mg·mL−1) | |||||
|---|---|---|---|---|---|---|
| 20 | 40 | 60 | 80 | 100 | ||
| EEP-A | 8.65 ± 0.83h | 10.33 ± 1.04g | 12.09 ± 0.95e | 13.66 ± 1.04d | 14.88 ± 2.05c | 0.8 |
| EEP-C | 11.79 ± 1.09e | 13.74 ± 0.04d | 14.28 ± 2.11d | 15.16 ± 0.94b | 16.62 ± 1.77a | 0.8 |
| 70% ethanol | 7.04 ± 0.14 | - | ||||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, X.; Pu, R.; Li, Y.; Wu, Z.; Li, C.; Miao, X.; Yang, W. Chemical Compositions of Propolis from China and the United States and their Antimicrobial Activities Against Penicillium notatum. Molecules 2019, 24, 3576. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules24193576
Xu X, Pu R, Li Y, Wu Z, Li C, Miao X, Yang W. Chemical Compositions of Propolis from China and the United States and their Antimicrobial Activities Against Penicillium notatum. Molecules. 2019; 24(19):3576. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules24193576
Chicago/Turabian StyleXu, Xiaolan, Ruixue Pu, Yujie Li, Zhenghong Wu, Chunxia Li, Xiaoqing Miao, and Wenchao Yang. 2019. "Chemical Compositions of Propolis from China and the United States and their Antimicrobial Activities Against Penicillium notatum" Molecules 24, no. 19: 3576. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules24193576