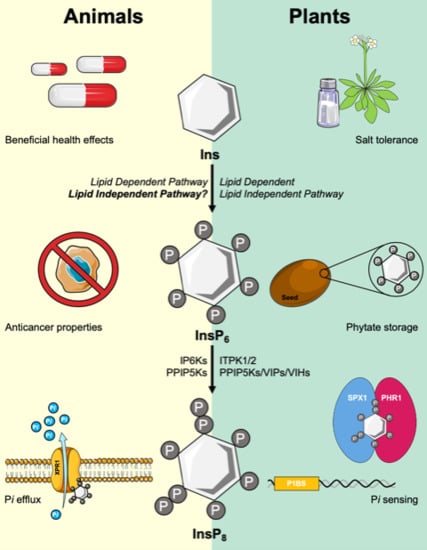
Inositol Pyrophosphate Pathways and Mechanisms: What Can We Learn from Plants?
Abstract
:1. Introduction
2. Myo-Inositol: The Signaling Scaffold and Precursor for the Lipid Independent Pathway
3. Is the Lipid-Independent InsP6 Synthetic Pathway Plant Specific?
4. Catalytic Flexibility in Plant and Human ITPKs
5. The PPIP5K/VIP/VIH Dual Domains are Arbiters of Biology
6. Do Human Signaling Pathways Share a Common Mechanism for Regulation by InsP8?
7. Concluding Remarks
Author Contributions
Funding
Conflicts of Interest
References
- Chakraborty, A.; Kim, S.; Snyder, S.H. Inositol pyrophosphates as mammalian cell signals. Sci. Signal. 2011, 4. [Google Scholar] [CrossRef] [Green Version]
- Hatch, A.J.; York, J.D. SnapShot: Inositol phosphates. Cell 2010, 143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Irvine, R.F. Inositide evolution-Towards turtle domination?: Inositide evolution. J. Physiol. 2005, 566, 295–300. [Google Scholar] [CrossRef] [PubMed]
- Shears, S.B.; Wang, H. Inositol phosphate kinases: Expanding the biological significance of the universal core of the protein kinase fold. Adv. Biol. Regul. 2019, 71, 118–127. [Google Scholar] [CrossRef] [PubMed]
- York, J.D. Regulation of nuclear processes by inositol polyphosphates. Biochim. Biophys. Acta 2006, 1761, 552–559. [Google Scholar] [CrossRef]
- Gillaspy, G.E. The cellular language of myo-inositol signaling. New Phytol. 2011, 192, 823–839. [Google Scholar] [CrossRef]
- Shears, S.B. Intimate connections: Inositol pyrophosphates at the interface of metabolic regulation and cell signaling. J. Cell. Physiol. 2018, 233, 1897–1912. [Google Scholar] [CrossRef]
- Raboy, V. Seeds for a better future: “low phytate” grains help to overcome malnutrition and reduce pollution. Trends Plant Sci. 2001, 6, 458–462. [Google Scholar] [CrossRef]
- White, P.J.; Veneklaas, E.J. Nature and nurture: The importance of seed phosphorus content. Plant Soil 2012, 357, 1–8. [Google Scholar] [CrossRef]
- Nagy, R.; Grob, H.; Weder, B.; Green, P.; Klein, M.; Frelet-Barrand, A.; Schjoerring, J.K.; Brearley, C.; Martinoia, E. The Arabidopsis ATP-binding cassette protein AtMRP5/AtABCC5 is a high affinity inositol hexakisphosphate transporterinvolved in guard cell signaling and phytate storage. J. Biol. Chem. 2009, 284, 33614–33622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Freed, C.; Adepoju, O.; Gillaspy, G. Can inositol pyrophosphates inform strategies for developing low phytate crops? Plants 2020, 9, 115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krinke, O.; Novotná, Z.; Valentová, O.; Martinec, J. Inositol trisphosphate receptor in higher plants: Is it real? J. Exp. Bot. 2007, 58, 361–376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michell, R.H. Inositol derivatives: Evolution and functions. Nat. Rev. Mol. Cell Biol. 2008, 9, 151–161. [Google Scholar] [CrossRef] [PubMed]
- Nelson, D.E.; Rammesmayer, G.; Bohnert, H.J. Regulation of cell-specific inositol metabolism and transport in plant salinity tolerance. Plant Cell 1998, 10, 753–764. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishitani, M.; Majumder, A.L.; Bornhouser, A.; Michalowski, C.B.; Jensen, R.G.; Bohnert, H.J. Coordinate transcriptional induction of myo-inositol metabolism during environmental stress. Plant J. 1996, 9, 537–548. [Google Scholar] [CrossRef] [PubMed]
- Carlomagno, G.; Unfer, V.; Roseff, S. The D-chiro-inositol paradox in the ovary. Fertil. Steril. 2011, 95, 2515–2516. [Google Scholar] [CrossRef]
- Galazis, N.; Galazi, M.; Atiomo, W. D-Chiro-inositol and its significance in polycystic ovary syndrome: A systematic review. Gynecol. Endocrinol. 2011, 27, 256–262. [Google Scholar] [CrossRef]
- Davis, A.; Christiansen, M.; Horowitz, J.F.; Klein, S.; Hellerstein, M.K.; Ostlund, R.E. Effect of pinitol treatment on insulin action in subjects with insulin resistance. Diabetes Care 2000, 23, 1000–1005. [Google Scholar] [CrossRef] [Green Version]
- Larner, J. D-chiro-inositol-its functional role in insulin action and its deficit in insulin resistance. Int. J. Exp. Diabetes Res. 2002, 3, 47–60. [Google Scholar] [CrossRef]
- Kang, M.-J.; Kim, J.-I.; Yoon, S.-Y.; Kim, J.C.; Cha, I.-J. Pinitol from soybeans reduces postprandial blood glucose in patients with type 2 diabetes mellitus. J. Med. Food 2006, 9, 182–186. [Google Scholar] [CrossRef]
- Nissen, P.M.; Nebel, C.; Oksbjerg, N.; Bertram, H.C. Metabolomics reveals relationship between plasma inositols and birth weight: Possible markers for fetal programming of type 2 diabetes. J. Biomed. Biotechnol. 2011, 2011. [Google Scholar] [CrossRef]
- Pintaudi, B.; Di Vieste, G.; Bonomo, M. The Effectiveness of Myo-Inositol and D-Chiro Inositol Treatment in Type 2 Diabetes. Int. J. Endocrinol. 2016, 2016. [Google Scholar] [CrossRef] [Green Version]
- Stevenson-Paulik, J.; Odom, A.R.; York, J.D. Molecular and biochemical characterization of two plant inositol polyphosphate 6-/3-/5-kinases. J. Biol. Chem. 2002, 277, 42711–42718. [Google Scholar] [CrossRef] [Green Version]
- Stevenson-Paulik, J.; Bastidas, R.J.; Chiou, S.T.; Frye, R.A.; York, J.D. Generation of phytate-free seeds in Arabidopsis through disruption of inositol polyphosphate kinases. Proc. Natl. Acad. Sci. USA 2005, 102, 12612–12617. [Google Scholar] [CrossRef] [Green Version]
- Berridge, M.J.; Irvine, R.F. Inositol phosphates and cell signalling. Nature 1989, 341, 197–205. [Google Scholar] [CrossRef]
- Saiardi, A.; Erdjument-Bromage, H.; Snowman, A.M.; Tempst, P.; Snyder, S.H. Synthesis of diphosphoinositol pentakisphosphate by a newly identified family of higher inositol polyphosphate kinases. Curr. Biol. 1999, 9, 1323–1326. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.I.; Tai, T.H. Identification of genes necessary for wild-type levels of seed phytic acid in Arabidopsis thaliana using a reverse genetics approach. Mol. Genet. Genom. 2011, 286, 119–133. [Google Scholar] [CrossRef]
- Shi, J.; Wang, H.; Hazebroek, J.; Ertl, D.S.; Harp, T. The maize low-phytic acid 3 encodes a myo-inositol kinase that plays a role in phytic acid biosynthesis in developing seeds. Plant J. 2005, 42, 708–719. [Google Scholar] [CrossRef]
- Funkhouser, E.A.; Loewus, F.A. Purification of myo-Inositol 1-Phosphate Synthase from Rice Cell Culture by Affinity Chromatography. Plant Physiol. 1975, 56, 786–790. [Google Scholar] [CrossRef] [Green Version]
- Desfougères, Y.; Wilson, M.S.C.; Laha, D.; Miller, G.J.; Saiardi, A. ITPK1 mediates the lipid-independent synthesis of inositol phosphates controlled by metabolism. Proc. Natl. Acad. Sci. USA 2019, 116, 24551–24561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilson, M.P.; Majerus, P.W. Characterization of a cDNA encoding Arabidopsis thaliana inositol 1,3,4-trisphosphate 5/6-kinase. Biochem. Biophys. Res. Commun. 1997, 232, 678–681. [Google Scholar] [CrossRef]
- Yang, X.; Shears, S.B. Multitasking in signal transduction by a promiscuous human Ins(3,4,5,6)P(4) 1-kinase/Ins(1,3,4)P(3) 5/6-kinase. Biochem. J. 2000, 351, 551–555. [Google Scholar] [CrossRef]
- Sweetman, D.; Johnson, S.; Caddick, S.E.K.; Hanke, D.E.; Brearley, C.A. Characterization of an Arabidopsis inositol 1,3,4,5,6-pentakisphosphate 2-kinase(AtIPK1). Biochem. J. 2006, 394, 95–103. [Google Scholar] [CrossRef] [Green Version]
- Verbsky, J.W.; Wilson, M.P.; Kisseleva, M.V.; Majerus, P.W.; Wente, S.R. The synthesis of inositol hexakisphosphate: Characterization of human inositol 1,3,4,5,6-pentakisphosphate 2-kinase. J. Biol. Chem. 2002, 277, 31857–31862. [Google Scholar] [CrossRef] [Green Version]
- Kuo, H.F.; Hsu, Y.Y.; Lin, W.C.; Chen, K.Y.; Munnik, T.; Brearley, C.A.; Chiou, T.J. Arabidopsis inositol phosphate kinases IPK1 and ITPK1 constitute a metabolic pathway in maintaining phosphate homeostasis. Plant J. 2018, 95, 613–630. [Google Scholar] [CrossRef] [Green Version]
- Draskovic, P.; Saiardi, A.; Bhandari, R.; Burton, A.; Ilc, G.; Kovacevic, M.; Snyder, S.H.; Podobnik, M. Inositol hexakisphosphate kinase products contain diphosphate and triphosphate groups. Chem. Biol. 2008, 15, 274–286. [Google Scholar] [CrossRef] [Green Version]
- Desai, M.; Rangarajan, P.; Donahue, J.L.; Williams, S.P.; Land, E.S.; Mandal, M.K.; Phillippy, B.Q.; Perera, I.Y.; Raboy, V.; Gillaspy, G.E. Two inositol hexakisphosphate kinases drive inositol pyrophosphate synthesis in plants. Plant J. 2014, 80, 642–653. [Google Scholar] [CrossRef]
- Sweetman, D.; Stavridou, I.; Johnson, S.; Green, P.; Caddick, S.E.K.; Brearley, C.A. Arabidopsis thaliana inositol 1,3,4-trisphosphate 5/6-kinase 4 (AtITPK4) is an outlier to a family of ATP-grasp fold proteins from Arabidopsis. FEBS Lett. 2007, 581, 4165–4171. [Google Scholar] [CrossRef] [Green Version]
- Qian, X.; Mitchell, J.; Wei, S.J.; Williams, J.; Petrovich, R.M.; Shears, S.B. The Ins(1,3,4)P3 5/6-kinase/Ins(3,4,5,6)P4 1-kinase is not a protein kinase. Biochem. J. 2005, 389, 389–395. [Google Scholar] [CrossRef] [Green Version]
- Adepoju, O.; Williams, S.P.; Craige, B.; Cridland, C.A.; Sharpe, A.K.; Brown, A.M.; Land, E.; Perera, I.Y.; Mena, D.; Sobrado, P.; et al. Inositol Trisphosphate Kinase and Diphosphoinositol Pentakisphosphate Kinase Enzymes Constitute the Inositol Pyrophosphate Synthesis Pathway in Plants. bioRxiv 2019, 724914. [Google Scholar] [CrossRef] [Green Version]
- Laha, D.; Parvin, N.; Hofer, A.; Giehl, R.F.H.; Fernandez-Rebollo, N.; Von Wirén, N.; Saiardi, A.; Jessen, H.J.; Schaaf, G. Arabidopsis ITPK1 and ITPK2 Have an Evolutionarily Conserved Phytic Acid Kinase Activity. ACS Chem. Biol. 2019, 14, 2127–2133. [Google Scholar] [CrossRef] [Green Version]
- Schell, M.J. Inositol trisphosphate 3-kinases: Focus on immune and neuronal signaling. Cell. Mol. Life Sci. 2010, 67, 1755–1778. [Google Scholar] [CrossRef]
- Wang, H.; Derose, E.F.; London, R.E.; Shears, S.B. IP6K structure and the molecular determinants of catalytic specificity in an inositol phosphate kinase family. Nat. Commun. 2014, 5. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Falck, J.R.; Hall, T.M.T.; Shears, S.B. Structural basis for an inositol pyrophosphate kinase surmounting phosphate crowding. Nat. Chem. Biol. 2012, 8, 111–116. [Google Scholar] [CrossRef] [Green Version]
- Mulugu, S.; Bai, W.; Fridy, P.C.; Bastidas, R.J.; Otto, J.C.; Dollins, D.E.; Haystead, T.A.; Ribeiro, A.A.; York, J.D. A conserved family of enzymes that phosphorylate inositol hexakisphosphate. Science 2007, 316, 106–109. [Google Scholar] [CrossRef]
- Choi, J.H.; Williams, J.; Cho, J.; Falck, J.R.; Shears, S.B. Purification, sequencing, and molecular identification of a mammalian PP-InsP5 kinase that is activated when cells are exposed to hyperosmotic stress. J. Biol. Chem. 2007, 282, 30763–30775. [Google Scholar] [CrossRef] [Green Version]
- Gokhale, N.A.; Zaremba, A.; Shears, S.B. Receptor-dependent compartmentalization of PPIP5K1, a kinase with a cryptic polyphosphoinositide binding domain. Biochem. J. 2011, 434, 415–426. [Google Scholar] [CrossRef] [Green Version]
- Scheffzek, K.; Welti, S. Pleckstrin homology (PH) like domains-versatile modules in protein-protein interaction platforms. FEBS Lett. 2012, 586, 2662–2673. [Google Scholar] [CrossRef] [Green Version]
- Nair, V.S.; Gu, C.; Janoshazi, A.K.; Jessen, H.J.; Wang, H.; Shears, S.B. Inositol pyrophosphate synthesis by diphosphoinositol pentakisphosphate kinase-1 is regulated by phosphatidylinositol(4,5)bisphosphate. Biosci. Rep. 2018, 38. [Google Scholar] [CrossRef] [Green Version]
- Luo, H.R.; Huang, Y.E.; Chen, J.C.; Saiardi, A.; Iijima, M.; Ye, K.; Huang, Y.; Nagata, E.; Devreotes, P.; Snyder, S.H. Inositol pyrophosphates mediate chemotaxis in Dictyostelium via pleckstrin homology domain-Ptdins(3,4,5)P3 interactions. Cell 2003, 114, 559–572. [Google Scholar] [CrossRef] [Green Version]
- Shears, S.B. A short historical perspective of methods in inositol phosphate research. In Methods in Molecular Biology; Miller, G., Ed.; Humana Press Inc.: Totowa, NJ, USA, 2020; Volume 2091, pp. 1–28. [Google Scholar]
- Randall, T.A.; Gu, C.; Li, X.; Wang, H.; Shears, S.B. A two-way switch for inositol pyrophosphate signaling: Evolutionary history and biological significance of a unique, bifunctional kinase/phosphatase. Adv. Biol. Regul. 2019, 75. [Google Scholar] [CrossRef] [PubMed]
- Machkalyan, G.; Trieu, P.; Pétrin, D.; Hébert, T.E.; Miller, G.J. PPIP5K1 interacts with the exocyst complex through a C-terminal intrinsically disordered domain and regulates cell motility. Cell. Signal. 2016, 28, 401–411. [Google Scholar] [CrossRef]
- Pisani, F.; Livermore, T.; Rose, G.; Chubb, J.R.; Gaspari, M.; Saiardi, A. Analysis of Dictyostelium discoideum inositol pyrophosphate metabolism by gel electrophoresis. PLoS ONE 2014, 9. [Google Scholar] [CrossRef] [Green Version]
- Dollins, D.E.; Bai, W.; Fridy, P.C.; Otto, J.C.; Neubauer, J.L.; Gattis, S.G.; Mehta, K.P.M.; York, J.D. Vip1 is a kinase and pyrophosphatase switch that regulates inositol diphosphate signaling. Proc. Natl. Acad. Sci. USA 2020, 117, 9356–9364. [Google Scholar] [CrossRef] [Green Version]
- Gu, C.; Nguyen, H.N.; Ganini, D.; Chen, Z.; Jessen, H.J.; Gu, Z.; Wang, H.; Shears, S.B. KO of 5-InsP7 kinase activity transforms the HCT116 colon cancer cell line into a hypermetabolic, growth-inhibited phenotype. Proc. Natl. Acad. Sci. USA 2017, 114, 11968–11973. [Google Scholar] [CrossRef] [Green Version]
- Pascual-Ortiz, M.; Saiardi, A.; Walla, E.; Jakopec, V.; Künzel, N.A.; Span, I.; Vangala, A.; Fleig, U. Asp1 Bifunctional Activity Modulates Spindle Function via Controlling Cellular Inositol Pyrophosphate Levels in Schizosaccharomyces pombe. Mol. Cell. Biol. 2018, 38. [Google Scholar] [CrossRef] [Green Version]
- Pöhlmann, J.; Risse, C.; Seidel, C.; Pohlmann, T.; Jakopec, V.; Walla, E.; Ramrath, P.; Takeshita, N.; Baumann, S.; Feldbrügge, M.; et al. The Vip1 Inositol Polyphosphate Kinase Family Regulates Polarized Growth and Modulates the Microtubule Cytoskeleton in Fungi. PLoS Genet. 2014, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azevedo, C.; Saiardi, A. Eukaryotic Phosphate Homeostasis: The Inositol Pyrophosphate Perspective. Trends Biochem. Sci. 2017, 42, 219–231. [Google Scholar] [CrossRef]
- Jung, J.-Y.; Ried, M.K.; Hothorn, M.; Poirier, Y. Control of plant phosphate homeostasis by inositol pyrophosphates and the SPX domain. Curr. Opin. Biotechnol. 2018, 49, 156–162. [Google Scholar] [CrossRef] [Green Version]
- Zhu, J.; Lau, K.; Puschmann, R.; Harmel, R.K.; Zhang, Y.; Pries, V.; Gaugler, P.; Broger, L.; Dutta, A.K.; Jessen, H.J.; et al. Two bifunctional inositol pyrophosphate kinases/phosphatases control plant phosphate homeostasis. eLife 2019, 8, 1–25. [Google Scholar] [CrossRef]
- Dong, J.; Ma, G.; Sui, L.; Wei, M.; Satheesh, V.; Zhang, R.; Ge, S.; Li, J.; Zhang, T.-E.; Wittwer, C.; et al. Inositol Pyrophosphate InsP8 Acts as an Intracellular Phosphate Signal in Arabidopsis. Mol. Plant 2019, 12, 1463–1473. [Google Scholar] [CrossRef] [PubMed]
- Kuo, H.F.; Chang, T.Y.; Chiang, S.F.; Wang, W.D.; Charng, Y.Y.; Chiou, T.J. Arabidopsis inositol pentakisphosphate 2-kinase, AtIPK1, is required for growth and modulates phosphate homeostasis at the transcriptional level. Plant J. 2014, 80, 503–515. [Google Scholar] [CrossRef] [PubMed]
- Wilson, M.S.; Henning, X.; Jessen, J.; Saiardi, A. The inositol hexakisphosphate kinases IP6K1 and-2 regulate human cellular phosphate homeostasis, including XPR1-mediated phosphate export Downloaded from. J. Biol. Chem 2019, 294, 11597–11608. [Google Scholar] [CrossRef] [Green Version]
- Wild, R.; Gerasimaite, R.; Jung, J.-Y.; Truffault, V.; Pavlovic, I.; Schmidt, A.; Saiardi, A.; Jessen, H.J.; Poirier, Y.; Hothorn, M.; et al. Control of eukaryotic phosphate homeostasis by inositol polyphosphate sensor domains. Science 2016, 352, 986–990. [Google Scholar] [CrossRef] [Green Version]
- Gerasimaite, R.; Pavlovic, I.; Capolicchio, S.; Hofer, A.; Schmidt, A.; Jessen, H.J.; Mayer, A. Inositol Pyrophosphate Specificity of the SPX-Dependent Polyphosphate Polymerase VTC. ACS Chem. Biol. 2017, 12, 648–653. [Google Scholar] [CrossRef] [PubMed]
- Secco, D.; Wang, C.; Arpat, B.A.; Wang, Z.; Poirier, Y.; Tyerman, S.D.; Wu, P.; Shou, H.; Whelan, J. The emerging importance of the SPX domain-containing proteins in phosphate homeostasis. New Phytol. 2012, 193, 842–851. [Google Scholar] [CrossRef] [Green Version]
- Secco, D.; Wang, C.; Shou, H.; Whelan, J. Phosphate homeostasis in the yeast Saccharomyces cerevisiae, the key role of the SPX domain-containing proteins. FEBS Lett. 2012, 586, 289–295. [Google Scholar] [CrossRef] [Green Version]
- Hürlimann, H.C.; Pinson, B.; Stadler-Waibel, M.; Zeeman, S.C.; Freimoser, F.M. The SPX domain of the yeast low-affinity phosphate transporter Pho90 regulates transport activity. EMBO Rep. 2009, 10, 1003–1008. [Google Scholar] [CrossRef]
- Zhou, Z.; Wang, Z.; Lv, Q.; Shi, J.; Zhong, Y.; Wu, P.; Mao, C. SPX proteins regulate Pi homeostasis and signaling in different subcellular level. Plant Signal. Behav. 2015, 10. [Google Scholar] [CrossRef]
- Puga, M.I.; Mateos, I.; Charukesi, R.; Wang, Z.; Franco-Zorrilla, J.M.; de Lorenzo, L.; Irigoyen, M.L.; Masiero, S.; Bustos, R.; Rodrigues, J.; et al. SPX1 is a phosphate-dependent inhibitor of PHOSPHATE STARVATION RESPONSE 1 in Arabidopsis. PNAS 2014, 111, 14947–14952. [Google Scholar] [CrossRef] [Green Version]
- Ried, M.K.; Wild, R.; Zhu, J.; Broger, L.; Harmel, R.K.; Hothorn, L.A.; Fiedler, D.; Hothorn, M. Inositol pyrophosphates promote the interaction of SPX domains with the coiled-coil motif of PHR transcription factors to regulate plant phosphate homeostasis. BioRxiv 2019. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Gu, C.; Hostachy, S.; Sahu, S.; Wittwer, C.; Jessen, H.J.; Fiedler, D.; Wang, H.; Shears, S.B. Control of XPR1-dependent cellular phosphate efflux by InsP 8 is an exemplar for functionally-exclusive inositol pyrophosphate signaling. Proc. Natl. Acad. Sci. USA 2020, 117. [Google Scholar] [CrossRef] [PubMed]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cridland, C.; Gillaspy, G. Inositol Pyrophosphate Pathways and Mechanisms: What Can We Learn from Plants? Molecules 2020, 25, 2789. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules25122789
Cridland C, Gillaspy G. Inositol Pyrophosphate Pathways and Mechanisms: What Can We Learn from Plants? Molecules. 2020; 25(12):2789. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules25122789
Chicago/Turabian StyleCridland, Caitlin, and Glenda Gillaspy. 2020. "Inositol Pyrophosphate Pathways and Mechanisms: What Can We Learn from Plants?" Molecules 25, no. 12: 2789. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules25122789