NMR-Based Metabolic Profiling of Edible Olives—Determination of Quality Parameters
Abstract
:1. Introduction
2. Results and Discussion
2.1. Sample Collection
2.2. Acquisition, Data Processing and Multivariate Analysis
Geographical Origin, Olive Cultivar, Processing Procedure
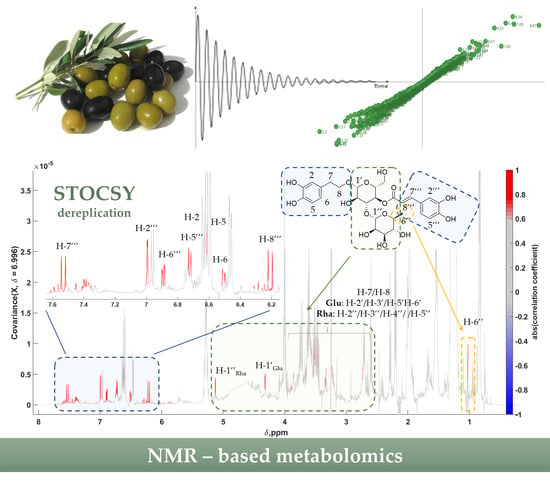
2.3. Statistical Total Correlation Spectroscopy (STOCSY) and Biomarker Identification
2.4. Quality and Authentication Assessment
3. Materials and Methods
3.1. Collection of Samples
3.2. Sample Preparation
3.2.1. Lyophilization
3.2.2. Extraction Protocol
3.3. NMR Analysis
3.3.1. Preparation of NMR Samples
3.3.2. NMR Experimental Parameters
3.4. Computational Processing and Multivariate Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- International Olive Council. Olives De Table - Table Olives - Consumption. Available online: https://www.internationaloliveoil.org/wp-content/uploads/2020/01/OT-W901-29-11-2019-C.pdf (accessed on 7 March 2020).
- International Olive Council. Key figures on the world market for table olives. Available online: https://www.internationaloliveoil.org/wp-content/uploads/2019/12/NEWSLETTER_144_ENGLISH.pdf (accessed on 7 March 2020).
- Ramírez, E.; Brenes, M.; De Castro, A.; Romero, C.; Medina, E. Oleuropein hydrolysis by lactic acid bacteria in natural green olives. LWT. - Food Sci. Technol. 2017, 78, 165–171. [Google Scholar]
- Johnson, R.L.; Mitchell, A.E. Reducing phenolics related to bitterness in table olives. J. Food Qual. 2018, 2018, 1–12. [Google Scholar] [CrossRef]
- Kailis, S.G. Olives. Encycl. Appl. Plant Sci. 2016, 3, 236–245. [Google Scholar]
- Ryan, D.; Robards, K. Phenolic compounds in olives. Analyst 1998, 123, 31–44. [Google Scholar] [CrossRef]
- Serreli, G.; Incani, A.; Atzeri, A.; Angioni, A.; Campus, M.; Cauli, E.; Zurru, R.; Deiana, M. Antioxidant effect of natural table olives phenolic extract against oxidative stress and membrane damage in enterocyte-like cells. J. Food Sci. 2017, 82, 380–385. [Google Scholar] [CrossRef]
- Gambino, C.M.; Accardi, G.; Aiello, A.; Candore, G.; Dara-Guccione, G.; Mirisola, M.; Procopio, A.; Taormina, G.; Caruso, C. Effect of extra virgin olive oil and table olives on the immuneinflammatory responses: Potential clinical applications. Endocr. Metab. Immune Disord. - Drug Targets 2017, 18, 14–22. [Google Scholar] [CrossRef]
- Accardi, G.; Aiello, A.; Gargano, V.; Gambino, C.M.; Caracappa, S.; Marineo, S.; Vesco, G.; Carru, C.; Zinellu, A.; Zarcone, M.; et al. Nutraceutical effects of table green olives: A pilot study with Nocellara del Belice olives. Immun. Ageing 2016, 13, 11–16. [Google Scholar] [CrossRef] [Green Version]
- Lozano-Mena, G.; Sánchez-González, M.; Juan, M.E.; Planas, J.M. Maslinic acid, a natural phytoalexin-type triterpene from olives - A promising nutraceutical? Molecules 2014, 19, 11538–11559. [Google Scholar] [CrossRef] [Green Version]
- Jiang, L.Q.; Takamura, H. Radical-scavenging compounds in olive fruits and their changes during table olive preparation. Appl. Mech. Mater. 2013, 295–298, 118–122. [Google Scholar] [CrossRef]
- Emilia Juan, M.; Wenzel, U.; Daniel, H.; Planas, J.M. Olive Fruit Extracts and HT-29 Human Colon Cancer Cells; Elsevier: Amsterdam, The Netherlands, 2010. [Google Scholar]
- Boskou, D. Olive fruit, table olives, and olive oil bioactive constituents. In Olive and Olive Oil Bioactive Constituents; Boskou, D., Ed.; AOCS PRESS: Urbana, IL, USA, 2015; pp. 1–30. [Google Scholar]
- Fiehn, O. Metabolomics - The link between genotypes and phenotypes. Plant Mol. Biol. 2002, 48, 155–171. [Google Scholar] [CrossRef]
- Durante, M.; Tufariello, M.; Tommasi, L.; Lenucci, M.S.; Bleve, G.; Mita, G. Evaluation of bioactive compounds in black table olives fermented with selected microbial starters. J. Sci. Food Agric. 2018, 98, 96–103. [Google Scholar] [CrossRef] [PubMed]
- Boskou, D.; Blekas, G.; Tsimidou, M. Phenolic compounds in olive oil and olives. Curr. Top. Nutraceutical Res. 2005, 3, 125–136. [Google Scholar]
- International Olive Council. Method: Sensory Analysis of Table Olives. Available online: https://www.internationaloliveoil.org/wp-content/uploads/2019/11/COI-OT-MO.-1-Rev.2-2011-Eng.pdf (accessed on 7 March 2020).
- Blekas, G.; Vassilakis, C.; Harizanis, C.; Tsimidou, M.; Boskou, D.G. Biophenols in table olives. J. Agric. Food Chem. 2002, 50, 3688–3692. [Google Scholar] [CrossRef] [PubMed]
- López-López, A.; Montaño, A.; Garrido-Fernández, A. Nutrient Profiles of Commercial Table Olives: Fatty Acids, Sterols, and Fatty Alcohols; Elsevier: Amsterdam, The Netherlands, 2010. [Google Scholar]
- Sahan, Y.; Cansev, A.; Gulen, H. Effect of processing techniques on antioxidative enzyme activities, antioxidant capacity, phenolic compounds, and fatty acids of table olives. Food Sci. Biotechnol. 2013, 22, 613–620. [Google Scholar] [CrossRef]
- European Union Commission Implementing Regulation (EU) No 668/2014. Off. J. Eur. Union 2014, L 179/36, 36–61.
- Luykx, D.M.A.M.; Van Ruth, S.M. An overview of analytical methods for determining the geographical origin of food products. Food Chem. 2008, 107, 897–911. [Google Scholar] [CrossRef]
- Böhme, K.; Calo-Mata, P.; Barros-Velázquez, J.; Ortea, I. Recent applications of omics-based technologies to main topics in food authentication. TrAC Trends Anal. Chem. 2019, 110, 221–232. [Google Scholar] [CrossRef]
- Versari, A.; Laurie, V.F.; Ricci, A.; Laghi, L.; Parpinello, G.P. Progress in authentication, typification and traceability of grapes and wines by chemometric approaches. Food Res. Int. 2014, 60, 2–18. [Google Scholar] [CrossRef]
- Steinmann, D.; Ganzera, M. Recent advances on HPLC/MS in medicinal plant analysis. J. Pharm. Biomed. Anal. 2011, 55, 744–757. [Google Scholar] [CrossRef]
- Soler, C.; Picó, Y. Recent trends in liquid chromatography-tandem mass spectrometry to determine pesticides and their metabolites in food. TrAC Trends Anal. Chem. 2007, 26, 103–115. [Google Scholar] [CrossRef]
- De Castro, A.; Sánchez, A.H.; López-López, A.; Cortés-Delgado, A.; Medina, E.; Montaño, A. Microbiota and metabolite profiling of spoiled spanish-style green table olives. Metabolites 2018, 8, 73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- López, A.; Montaño, A.; García, P.; Garrido, A. Fatty acid profile of table olives and its multivariate characterization using unsupervised (PCA) and supervised (DA) chemometrics. J. Agric. Food Chem. 2006, 54, 6747–6753. [Google Scholar] [CrossRef] [PubMed]
- López-López, A.; Jiménez-Araujo, A.; García-García, P.; Garrido-Fernández, A. Multivariate analysis for the evaluation of fiber, sugars, and organic acids in commercial presentations of table olives. J. Agric. Food Chem. 2007, 55, 10803–10811. [Google Scholar] [CrossRef] [PubMed]
- López, A.; García, P.; Garrido, A. Multivariate characterization of table olives according to their mineral nutrient composition. Food Chem. 2008, 106, 369–378. [Google Scholar] [CrossRef]
- Vallone, M.; Alleri, M.; Bono, F.; Catania, P. Use of a portable VIS NIR device to predict table olives quality. Chem. Eng. Trans. 2019, 75, 79–84. [Google Scholar]
- Boskou, G. Antioxidant Capacity and Phenolic Profile of Table Olives from the Greek Market; Elsevier: Amsterdam, The Netherlands, 2010. [Google Scholar]
- Gougoulias, N.; Giurgiulescu, L.; Vagelas, I.; Wogiatzi, E.; Ntalla, M.N. Changes in total phenol content and antioxidant activity of greek table olive cultivar amfissis during maturation. Stud. Univ. Babes-Bolyai Chem. 2017, 62, 387–396. [Google Scholar] [CrossRef]
- Mitsopoulos, G.; Papageorgiou, V.; Komaitis, M.; Hagidimitriou, M. Total phenolic content, phenolic profile and antioxidant activity of leaves and drupes in major Greek olive varieties. Not. Bot. Horti. Agrobot. 2016, 44, 155–161. [Google Scholar] [CrossRef] [Green Version]
- Mastralexi, A.; Mantzouridou, F.T.; Tsimidou, M.Z. Evolution of safety and other quality parameters of the greek pdo table olives “prasines elies chalkidikis” during industrial scale processing and storage. Eur. J. Lipid Sci. Technol. 2019, 121, 1800171. [Google Scholar] [CrossRef]
- Panagou, E.Z.; Tassou, C.C.; Skandamis, P.N. Physicochemical, microbiological, and organoleptic profiles of Greek table olives from retail outlets. J. Food Prot. 2006, 69, 1732–1738. [Google Scholar] [CrossRef]
- Alexandraki, V.; Georgalaki, M.; Papadimitriou, K.; Anastasiou, R.; Zoumpopoulou, G.; Chatzipavlidis, I.; Papadelli, M.; Vallis, N.; Moschochoritis, K.; Tsakalidou, E. Determination of triterpenic acids in natural and alkaline-treated Greek table olives throughout the fermentation process. LWT—Food Sci. Technol. 2014, 58, 609–613. [Google Scholar] [CrossRef]
- Zoidou, E.; Melliou, E.; Gikas, E.; Tsarbopoulos, A.; Magiatis, P.; Skaltsounis, A.L. Identification of throuba thassos, a traditional Greek table olive variety, as a nutritional rich source of oleuropein. J. Agric. Food Chem. 2010, 58, 46–50. [Google Scholar] [CrossRef] [PubMed]
- Rotondo, A.; Mannina, L.; Salvo, A. Multiple assignment recovered analysis (MARA) NMR for a direct food labeling: The case study of olive oils. Food Anal. Methods 2019, 12, 1238–1245. [Google Scholar] [CrossRef]
- Spiteri, M.; Jamin, E.; Thomas, F.; Rebours, A.; Lees, M.; Rogers, K.M.; Rutledge, D.N. Fast and global authenticity screening of honey using 1H-NMR profiling. Food Chem. 2015, 189, 60–66. [Google Scholar] [CrossRef] [PubMed]
- Jakes, W.; Gerdova, A.; Defernez, M.; Watson, A.D.; McCallum, C.; Limer, E.; Colquhoun, I.J.; Williamson, D.C.; Kemsley, E.K. Authentication of beef versus horse meat using 60 MHz 1H NMR spectroscopy. Food Chem. 2015, 175, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Halabalaki, M.; Vougogiannopoulou, K.; Mikros, E.; Skaltsounis, A.L. Recent advances and new strategies in the NMR-based identification of natural products. Curr. Opin. Biotechnol. 2014, 25, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Cloarec, O.; Dumas, M.-E.; Craig, A.; Barton, R.H.; Trygg, J.; Hudson, J.; Blancher, C.; Gauguier, D.; Lindon, J.C.; Holmes, E.; et al. Statistical total correlation spectroscopy: An exploratory approach for latent biomarker identification from metabolic 1 H NMR data sets. Anal. Chem. 2005, 77, 1282–1289. [Google Scholar] [CrossRef]
- Boka, V.I.; Stathopoulou, K.; Benaki, D.; Gikas, E.; Aligiannis, N.; Mikros, E.; Skaltsounis, A.L. Could multivariate statistics exploit HPTLC and NMR data to reveal bioactive compounds? The case of Paeonia mascula. Phytochem. Lett. 2017, 20, 379–385. [Google Scholar] [CrossRef]
- Yilmaz, A.; Nyberg, N.T.; Jaroszewski, J.W. Extraction of alkaloids for NMR-based profiling: Exploratory analysis of an archaic Cinchona bark collection. Planta Med. 2012, 78, 1885–1890. [Google Scholar] [CrossRef] [Green Version]
- Guldbrandsen, N.; Kostidis, S.; Schäfer, H.; De Mieri, M.; Spraul, M.; Skaltsounis, A.L.; Mikros, E.; Hamburger, M. NMR-based metabolomic study on isatis tinctoria: Comparison of different accessions, harvesting dates, and the effect of repeated harvesting. J. Nat. Prod. 2015, 78, 977–986. [Google Scholar] [CrossRef]
- Freire, R.T.; Bero, J.; Beaufay, C.; Selegato, D.M.; Coqueiro, A.; Choi, Y.H.; Quetin-Leclercq, J. Identification of antiplasmodial triterpenes from Keetia species using NMR-based metabolic profiling. Metabolomics 2019, 15, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Kang, J.; Choi, M.Y.; Kang, S.; Hyuk, N.K.; Wen, H.; Chang, H.L.; Park, M.; Wiklund, S.; Hyo, J.K.; Sung, W.K.; et al. Application of a 1H nuclear magnetic resonance (NMR) metabolomics approach combined with orthogonal projections to latent structure-discriminant analysis as an efficient tool for discriminating between Korean and Chinese herbal medicines. J. Agric. Food Chem. 2008, 56, 11589–11595. [Google Scholar] [CrossRef] [PubMed]
- Lopez, J.M.; Cabrera, R.; Maruenda, H. Ultra-clean pure shift 1H-NMR applied to metabolomics profiling. Sci. Rep. 2019, 9, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Bo, Y.; Feng, J.; Xu, J.J.; Huang, Y.; Cai, H.; Cui, X.; Dong, J.; Ding, S.; Chen, Z. High-resolution pure shift NMR spectroscopy offers better metabolite discrimination in food quality analysis. Food Res. Int. 2019, 125, 108574. [Google Scholar] [CrossRef] [PubMed]
- Kew, W.; Goodall, I.; Uhrín, D. Analysis of scotch whisky by 1H NMR and chemometrics yields insight into its complex chemistry. Food Chem. 2019, 298, 125052. [Google Scholar] [CrossRef] [PubMed]
- Ghanbari, R.; Anwar, F.; Alkharfy, K.M.; Gilani, A.H.; Saari, N. Valuable nutrients and functional bioactives in different parts of olive (Olea europaea L.)-A review. Int. J. Mol. Sci. 2012, 13, 3291–3340. [Google Scholar] [CrossRef] [PubMed]
- Evangelou, E.; Kiritsakis, K.; Sakellaropoulos, N.; Kiritsakis, A. Table olives production, postharvest processing, and nutritional qualities. In Handbook of Vegetables and Vegetable Processing; Wiley: Hoboken, NJ, USA, 2018; pp. 727–744. [Google Scholar]
- Rotondo, A.; Salvo, A.; Gallo, V.; Rastrelli, L.; Dugo, G. Quick unreferenced NMR quantification of Squalene in vegetable oils. Eur. J. Lipid Sci. Technol. 2017, 119, 1700151. [Google Scholar] [CrossRef] [Green Version]
- Danezis, G.P.; Tsagkaris, A.S.; Camin, F.; Brusic, V.; Georgiou, C.A. Food authentication: Techniques, trends & emerging approaches. TrAC Trends Anal. Chem. 2016, 85, 123–132. [Google Scholar]
- Sugiyama, Y.; Ito, Y.; Suzuki, M.; Hirota, A. Indole derivatives from a marine sponge-derived yeast as DPPH radical scavengers. J. Nat. Prod. 2009, 72, 2069–2071. [Google Scholar] [CrossRef]
- Kalampaliki, A.D.; Giannouli, V.; Skaltsounis, A.L.; Kostakis, I.K. A three-step, gram-scale synthesis of hydroxytyrosol, hydroxytyrosol acetate, and 3,4-dihydroxyphenylglycol. Molecules 2019, 24, 3239. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Zhou, S.; Xu, G.; Gao, Y. Interference of phenylethanoid glycosides from cistanche tubulosa with the MTT assay. Molecules 2015, 20, 8060–8071. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Li, X.; Zhang, S.; Lu, W.; Liao, S.; Liu, X.; Shan, L.; Shen, X.; Jiang, H.; Zhang, W.; et al. Natural products as a gold mine for selective matrix metalloproteinases inhibitors. Bioorg. Med. Chem. 2012, 20, 4164–4171. [Google Scholar] [CrossRef] [PubMed]
- Tasnuva, S.T.; Qamar, U.A.; Ghafoor, K.; Sahena, F.; Jahurul, M.H.A.; Rukshana, H.; Juliana, M.J.; Al-juhaimi, F.Y.; Jalifah, L.; Jalal, K.C.A.; et al. α -glucosidase inhibitors isolated from Mimosa pudica L. Nat. Prod. Res. 2019, 33, 1495–1499. [Google Scholar] [CrossRef] [PubMed]
- Lima, R.D.C.L.; Kongstad, K.T.; Kato, L.; Das Silva, M.J.; Franzyk, H.; Staerk, D. High-resolution PTP1B inhibition profiling combined with HPLC-HRMS-SPE-NMR for identification of PTP1B inhibitors from Miconia albicans. Molecules 2018, 23, 1755. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Albergamo, A.; Rotondo, A.; Salvo, A.; Pellizzeri, V.; Bua, D.G.; Maggio, A.; Cicero, N.; Dugo, G.; Albergamo, A.; Rotondo, A.; et al. Metabolite and mineral profiling of “ Violetto di Niscemi ” and “ Spinoso di Menfi ” globe artichokes by H-NMR and ICP-MS. Nat. Prod. Res. 2017, 31, 990–999. [Google Scholar] [CrossRef]
- Monguchi, Y.; Ichikawa, T.; Nozaki, K.; Kihara, K.; Yamada, Y.; Miyake, Y.; Sawama, Y.; Sajiki, H. Development of chelate resin-supported palladium catalysts for chemoselective hydrogenation. Tetrahedron 2015, 71, 6499–6505. [Google Scholar] [CrossRef]
- Gottlieb, H.E.; Kotlyar, V.; Nudelman, A. NMR chemical shifts of common laboratory solvents as trace impurities. J. Org. Chem. 1997, 62, 7512–7515. [Google Scholar] [CrossRef]
- Ruiz-Aracama, A.; Goicoechea, E.; Guillén, M.D. Direct study of minor extra-virgin olive oil components without any sample modification. 1H NMR multisupression experiment: A powerful tool. Food Chem. 2017, 228, 301–314. [Google Scholar] [CrossRef]
- Rathnayake, G.R.N.; Kumar, N.S.; Jayasinghe, L.; Araya, H.; Fujimoto, Y. Chemical investigation of metabolites produced by an endophytic fungi Phialemonium curvatum from the leaves of Passiflora edulis. Nat. Prod. Res. 2018, 32, 2483–2486. [Google Scholar] [CrossRef]
- Dugo, G.; Rotondo, A.; Mallamace, D.; Cicero, N.; Salvo, A.; Rotondo, E.; Corsaro, C. Enhanced detection of aldehydes in extra-virgin olive oil by means of band selective NMR spectroscopy. Phys. A Stat. Mech. Its Appl. 2015, 420, 258–264. [Google Scholar] [CrossRef]
- Klikarová, J.; Rotondo, A.; Cacciola, F.; Česlová, L.; Dugo, P.; Mondello, L.; Rigano, F. The phenolic fraction of italian extra virgin olive oils: Elucidation through combined liquid chromatography and NMR approaches. Food Anal. Methods 2019, 12, 1759–1770. [Google Scholar] [CrossRef]
- Moreno-González, R.; Juan, M.E.; Planas, J.M. Table olive polyphenols: A simultaneous determination by liquid chromatography–mass spectrometry. J. Chromatogr. A 2019, 1609, 460434. [Google Scholar] [CrossRef] [PubMed]
- Kanakis, P.; Termentzi, A.; Michel, T.; Gikas, E.; Halabalaki, M.; Skaltsounis, A.L. From olive drupes to olive oil. An HPLC-orbitrap-based qualitative and quantitative exploration of olive key metabolites. Planta Med. 2013, 79, 1576–1587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cabrera-Bañegil, M.; Schaide, T.; Manzano, R.; Delgado-Adámez, J.; Durán-Merás, I.; Martín-Vertedor, D. Optimization and validation of a rapid liquid chromatography method for determination of the main polyphenolic compounds in table olives and in olive paste. Food Chem. 2017, 233, 164–173. [Google Scholar] [CrossRef]
- Ayeleso, T.B. Oleanolic acid and its derivatives: Biological activities and therapeutic potential in chronic diseases. Molecules 2017, 22, 1915. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pasqualone, A.; Nasti, R.; Montemurro, C.; Gomes, T. Effect of natural-style processing on the oxidative and hydrolytic degradation of the lipid fraction of table olives. Food Control 2014, 37, 99–103. [Google Scholar] [CrossRef]
- Alves, E.; Melo, T.; Barros, M.P.; Ros, M.R.M.; Domingues, P. Lipidomic Profiling of the Olive (Olea europaea L.) fruit towards its valorisation as a functional food: In-depth identification of triacylglycerols and polar. Molecules 2019, 24, 2555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsantili, E. Quality attributes and their relations in fresh black ripe “Kalamon” olives (Olea europaea L.) for table use - phenolic compounds and total antioxidant capacity. Int. J. Food Sci. Technol. 2014, 49, 657–665. [Google Scholar] [CrossRef]
- Gentile, L.; Uccella, N.A. Selected bioactives from callus cultures of olives (Olea europaea L. Var. Coratina) by LC-MS. Food Res. Int. 2014, 55, 128–136. [Google Scholar] [CrossRef]
- Bastoni, L.; Bianco, A.; Piccioni, F.; Uccella, N. Biophenolic profile in olives by nuclear magnetic resonance. Food Chem. 2001, 73, 145–151. [Google Scholar] [CrossRef]
- Bouaziz, M.; Jemai, H.; Khabou, W.; Sayadi, S. Oil content, phenolic profiling and antioxidant potential of Tunisian olive drupes. J. Sci. Food Agric. 2010, 90, 1750–1758. [Google Scholar] [CrossRef]
- Brenes, M.; Medina, E.; García, A.; Romero, C.; De Castro, A. Olives and Olive Oil Compounds Active Against Pathogenic Microorganisms; Elsevier: Amsterdam, The Netherlands, 2010. [Google Scholar]
- Emília Juan, M.; Wenzel, U.; Daniel, H.; Planas, J.M. Cancer Chemopreventive Activity of Hydroxytyrosol: A Natural Antioxidant from Olives and Olive Oil; Elsevier: Amsterdam, The Netherlands, 2010. [Google Scholar]
- Scientific Opinion on the substantiation of health claims related to polyphenols in olive and protection of LDL particles from oxidative damage (ID 1333, 1638, 1639, 1696, 2865), maintenance of normal blood HDL-cholesterol concentrations (ID 1639), mainte. Efsa J. 2011, 9, 2033. [CrossRef]
- Montano, A.; Sanchez, A.H.; Casado, F.J.; De Castro, A.; Rejano, L. Chemical profile of industrially fermented green olives of different varieties. Food Chem. 2003, 82, 297–302. [Google Scholar] [CrossRef]
- Sabatini, N.; Marsilio, V. Volatile compounds in table olives (Olea Europaea L., Nocellara del Belice cultivar). Food Chem. 2008, 107, 1522–1528. [Google Scholar] [CrossRef]
- International Olive Council. Determination of Biophenols in Oive Oils by HPLC. 2017, pp. 1–8. Available online: https://www.internationaloliveoil.org/wp-content/uploads/2019/11/COI-T.20-Doc.-No-29-Rev-1-2017.pdf (accessed on 7 March 2020).
- Eriksson, L.; Byrne, T.; Johansson, E.; Trygg, J.; Wikström, C. Multi- and Megavariate Data Analysis Basic Principles and Applications, 3rd ed.; Umetrics Academy: SAN Jose, CA, USA, 2013. [Google Scholar]
- Xia, J.; Wishart, D.S. Using metaboanalyst 3.0 for comprehensive metabolomics data analysis. Curr. Protoc. Bioinforma. 2016, 2016, 14.10.1–14.10.91. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: All analyzed samples are available for further evaluation in the Division of Pharmacognosy and Natural Products Chemistry, Faculty of Pharmacy, National and Kapodistrian University of Athens. |









| Region | Subregion | Variety | Processing Type | Sample Number | |
|---|---|---|---|---|---|
| 1 | Makedonia | Chalkidiki | Chalkidikis | Spanish (green) | 9 |
| 2 | Kavala | Chalkidikis | 6 | ||
| 3 | Sterea Ellada | Magnesia | Konservolia | Greek (black) | 6 |
| 4 | Fthiotida | Konservolia | 9 | ||
| 5 | Fthiotida | Kalamon | 1 | ||
| 6 | Aitoloakarnania | Kalamon | 14 | ||
| 7 | Peloponnese | Messinia | Kalamon | 10 | |
| 8 | Lakonia | Kalamon | 5 |
| No | Compound | δ1H (Multiplicity, J in Hz, Assignment) | Ref. |
|---|---|---|---|
| 1 | Tyr | 6.96 (d, J = 8.5 Hz, H-2, H-6), 6.64 (d, J = 8.5 Hz, H-3, H-5), 3.62 (t, J = 7.2 Hz, H-8), 2.65 (t, J = 7.2 Hz, H-7) | [56] |
| 2 | HT | 6.62 (d, J = 8.0 Hz, H-5), 6.59 (d, J = 1.9 Hz, H-2), 6.46 (dd, J = 1.9/8.0 Hz, H-6), 3.61 (t, J = 7.2 Hz, H-8), 2.60 (t, J = 7.2 Hz, H-7) | [57] |
| 3 | Ver | 7.53 (d, J = 15.9 Hz, H-7′’’), 7.00 (d, J = 2.0 Hz, H-2′’’), 6.89 (dd, J = 2.0/8.2 Hz, H6′’’), 6.72 (d, J = 8.2 Hz, H-5′’’), 6.63 (m, H-2), 6.61 (m, H-5), 6.50 (dd, J = 2.0/8.0 Hz, H-6), 6.21 (d, J = 15.9 Hz, H-8′’’), 5.13 (d, J = 1.7 Hz, H-1′’), 4.85 (m, H-4′), 4.32 (d, J = 8.0 Hz, H-1′), 3.98-2.73(m, H-7, H-8, H-2′, H-3′, H-5′, H-6′, H-2′’, H-3′’, H-4′’, H-5′’), 1.03 (d, J = 6.2 Hz, H-6′’) | [58] |
| 4 | Lut | 7.32 (dd, J = 2.2/9.0 Hz, H-6′), 7.32 (d, J = 2.2 Hz, H-2′), 6.85 (d, J = 9.0 Hz, H-5′), 6.48 (s, H-3), 6.39 (d, J = 2.1 Hz, H-8), 6.15 (d, J = 2.1 Hz, H-6) | [59] |
| 5 | Quer | 7.67 (d, J = 2.2 Hz, H-2′), 7.57 (dd, J = 2.2/8.5 Hz, H-6′), 6.83 (d, J = 8.5 Hz, H-5′), 6.34 (d, J = 2.0 Hz, H-8), 6.13 (d, J = 2.0 Hz, H-6) | [60] |
| 6 | MA | 5.19 (brt, J = 3.6 Hz, H-12), 3.55 (m, H-2), 2.85 (d, J = 9.6 Hz, H-3), 2.80 (dd, J = 4.1/14.1 Hz, H-18), 1.10 (s, H-27), 0.95 (s, H-24), 0.94 (s, H-23), 0.88 (s, H-30), 0.85 (s, H-29), 0.76 (s, H-26), 0.75 (s, H-25) | [61] |
| 7 | OA | 5.18 (brt, J = 3.5 Hz, H-12), 3.08 (dd, J = 4.9/11.7 Hz, H-3), 2.79 (dd, J = 4.1/14.3 Hz, H-18), 1.09 (s, H-27), 0,91 (s, H-23), 0.88 (s, H-25, H-30), 0.85 (s, H-29), 0.76 (s, H-26), 0.71 (s, H-24) | [61] |
| 8 | SA | 2.46 (s, H-2, H-3) | [62] |
| 9 | LA | 4.04 (q, J = 6.9 Hz, H-2), 1.29 (d, J = 6.9 Hz, H-3) | [62] |
| 10 | PA | 2.13 (q, J = 7.6 Hz, H-2), 1.03 (t, J = 7.6 Hz, H-3) | [63] |
| 11 | AA | 1.85 (s, H-2) | [64] |
| 12 | FA | 8.45 (s) | [64] |
| 13 | TAGs | 5.28 (m, olefinic protons), 5,25 (m, glyceryl group, H-2), 4.08 (dd, J = 4.4/11.4 Hz, glyceryl group, H-1a, H-3a), 4.01 (dd, 6.2/11.4 Hz, glyceryl group, H-1b, H-3b), 2.20 (t, J = 7.5 Hz, acyl groups, -OCO-CH2-), 2.02-1.95 (m, acyl groups, allylic protons), 1.54 (m, acyl groups, -OCO-CH2-CH2-), 1.34-1.18 (m, acyl groups, -(CH2)n-), 0.83 (t, 7.0, saturated/ monosaturated methyl group) | [65] |
| 14 | Lin* | 5.28 (m, olefinic protons), 5,25 (m, glyceryl group, H-2), 4.08 (dd, J = 4.4/11.4 Hz, glyceryl group, H-1a, H-3a), 4.01 (dd, 6.2/11.4 Hz, glyceryl group, H-1b, H-3b), 2.71 (t, J = 6.7 Hz, H-11, bis-allylic proton), 2.20 (t, J = 7.5 Hz, acyl groups, -OCO-CH2-), 2.02-1.95 (m, acyl groups, allylic protons), 1.54 (m, acyl groups, -OCO-CH2-CH2-), 1.34-1.18 (m, acyl groups, -(CH2)n-), 0.84 (t, J = 7.0 Hz, H-18) | [65] |
| 15 | G** | 3.59 (m, H-2), 3.53 (dd, J = 4.8/11.2 Hz, H-1a, H-3a), 3.46 (dd, J = 6.0/11.1 Hz, H-1b, H-3b) | [66] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Beteinakis, S.; Papachristodoulou, A.; Gogou, G.; Katsikis, S.; Mikros, E.; Halabalaki, M. NMR-Based Metabolic Profiling of Edible Olives—Determination of Quality Parameters. Molecules 2020, 25, 3339. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules25153339
Beteinakis S, Papachristodoulou A, Gogou G, Katsikis S, Mikros E, Halabalaki M. NMR-Based Metabolic Profiling of Edible Olives—Determination of Quality Parameters. Molecules. 2020; 25(15):3339. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules25153339
Chicago/Turabian StyleBeteinakis, Stavros, Anastasia Papachristodoulou, Georgia Gogou, Sotirios Katsikis, Emmanuel Mikros, and Maria Halabalaki. 2020. "NMR-Based Metabolic Profiling of Edible Olives—Determination of Quality Parameters" Molecules 25, no. 15: 3339. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules25153339