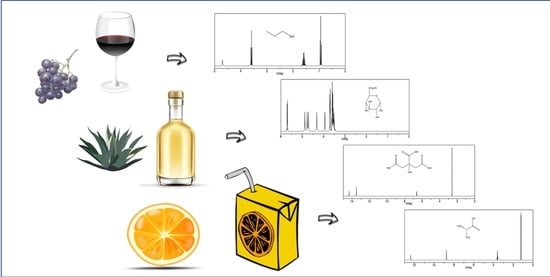
Nuclear Magnetic Resonance Metabolomics with Double Pulsed-Field-Gradient Echo and Automatized Solvent Suppression Spectroscopy for Multivariate Data Matrix Applied in Novel Wine and Juice Discriminant Analysis
Abstract
:1. Introduction
2. Results
2.1. Wines’ NMR Outliers with Double Pulsed-Field-Gradient Echo
2.2. Juice NMR Outliers with Automatized {1Hwater_presat NMR} -1D Single-Pulse NOESY
2.3. Multivariate Statistical Analysis
3. Discussion
3.1. Discriminant Analysis of Non-Saccharomyces Large-Scale Alcohol Reductions with Double Pulsed-Field-Gradient Echo NMR Metabolomics
3.2. Discriminant Analysis between “Type of Fruit” and “Type of Juice” in Different 100% Apple, Orange, and Apricot Beverages with Automatized {1Hwater_presat NMR} Metabolomics
4. Materials and Methods
4.1. Materials
4.1.1. Cabernet Sauvignon Wines Fermented at Different Large-Scale Conditions
4.1.2. Apple, Orange, and Apricot Juice Rroducts: (a) Made from Concentrate, (b) Not from Concentrate, (c) Nectars, and (d) Purees
4.2. Methods
4.2.1. NMR Spectroscopy of the Cabernet Sauvignon Wine Samples Fermented with Saccharomyces Cerevisiae, Candida Zemplinina, and Saccharomyces Bayanus Ex Uvarum Yeast Strains
4.2.2. NMR Spectroscopy of Apple, Orange, and Apricot 100% Juices Made from Concentrate, Not from Concentrate, Nectars, and Purees
4.2.3. H-NMR Postprocessing and Multivariate Statistical Analysis (MSA)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Pustjens, A.M.; Weesepoel, Y.; van Ruth, S.M. 1-Food Fraud and Authenticity: Emerging Issues and Future Trends, In Innovation and Future Trends in Food Manufacturing and Supply Chain Technologies; Leadley, C.E., Ed.; Woodhead Publishing: Cambridge, England, 2016; pp. 3–20. [Google Scholar] [CrossRef]
- Sobolev, A.P.; Thomas, F.; Donarski, J.; Ingallina, C.; Circi, S.; Marincola, F.C.; Capitani, D.; Mannina, L. Use of NMR applications to tackle future food fraud issues. Trends Food. Sci. Technol. 2019, 91, 347–353. [Google Scholar] [CrossRef] [Green Version]
- de Moura Ribeiro, M.V.; Boralle, N.; Pezza, H.R.; Pezza, L.; Toci, A.T. Authenticity of roasted coffee using 1H NMR spectroscopy. J. Food Compos. Anal. 2017, 57, 24–30. [Google Scholar] [CrossRef] [Green Version]
- Milani, M.I.; Rossini, E.L.; Catelani, T.A.; Pezza, L.; Toci, A.T.; Pezza, H.R. Authentication of roasted and ground coffee samples containing multiple adulterants using NMR and a chemometric approach. Food Control 2020, 112, 107104. [Google Scholar] [CrossRef]
- Gilard, V.; Balayssac, S.; Tinaugus, A.; Martins, N.; Martino, R.; Malet-Martino, M. Detection, identification and quantification by 1H NMR of adulterants in 150 herbal dietary supplements marketed for improving sexual performance. J. Pharmaceut. Biomed. 2015, 102, 476–493. [Google Scholar] [CrossRef]
- Hachem, R.; Assemat, G.; Martins, N.; Balayssac, S.; Gilard, V.; Martino, R.; Malet-Martino, M. Proton NMR for detection, identification and quantification of adulterants in 160 herbal food supplements marketed for weight loss. J. Pharmaceut. Biomed. 2016, 124, 34–47. [Google Scholar] [CrossRef]
- Shi, T.; Zhu, M.; Chen, Y.; Yan, X.; Chen, Q.; Wu, X.; Lin, J.; Xie, M. 1H NMR combined with chemometrics for the rapid detection of adulteration in camellia oils. Food Chem. 2018, 242, 308–315. [Google Scholar] [CrossRef]
- Bo, Y.; Feng, J.; Xu, J.; Huang, Y.; Cai, H.; Cui, X.; Dong, J.; Ding, S.; Chen, Z. High-resolution pure shift NMR spectroscopy offers better metabolite discrimination in food quality analysis. Food Res. Int. 2019, 125, 108574. [Google Scholar] [CrossRef] [PubMed]
- Kuballa, T.; Hausler, T.; Okaru, A.O.; Neufeld, M.; Abuga, K.O.; Kibwage, I.O.; Rehm, J.; Luy, B.; Walch, S.G.; Lachenmeier, D.W. Detection of counterfeit brand spirits using 1H NMR fingerprints in comparison to sensory analysis. Food Chem. 2018, 245, 112–118. [Google Scholar] [CrossRef] [PubMed]
- Ciepielowski, G.; Pacholczyk-Sienicka, B.; Frączek, T.; Klajman, K.; Paneth, P.; Albrecht, Ł. Comparison of quantitative NMR and IRMS for the authentication of ‘Polish Vodka’. J. Sci. Food Agric. 2019, 99, 263–268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santos, P.M.; Pereira-Filho, E.R.; Colnago, L.A. Detection and quantification of milk adulteration using time domain nuclear magnetic resonance (TD-NMR). Microchem. J. 2016, 124, 15–19. [Google Scholar] [CrossRef]
- Li, Q.; Yu, Z.; Zhu, D.; Meng, X.; Pang, X.; Liu, Y.; Frew, R.; Chen, H.; Chen, G. The application of NMR-based milk metabolite analysis in milk authenticity identification. J. Sci. Food Agric. 2017, 97, 2875–2882. [Google Scholar] [CrossRef] [PubMed]
- Hong, E.; Lee, S.Y.; Jeong, J.Y.; Park, J.M.; Kim, B.H.; Kwon, K.; Chun, H.S. Modern analytical methods for the detection of food fraud and adulteration by food category. J. Sci. Food Agric. 2017, 97, 3877–3896. [Google Scholar] [CrossRef]
- Schievano, E.; Sbrizza, M.; Zuccato, V.; Piana, L.; Tessari, M. NMR carbohydrate profile in tracing acacia honey authenticity. Food Chem. 2020, 309, 125788. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Liu, Y.; Liu, H.; Zheng, X.; Shen, G.; Feng, J. Compositional identification and authentication of Chinese honeys by 1H NMR combined with multivariate analysis. Food Res. Int. 2020, 130, 108936. [Google Scholar] [CrossRef] [PubMed]
- Lachenmeier, D.W. Advances in the detection of the adulteration of alcoholic beverages including unrecorded alcohol. In Advances in Food Authenticity Testing; Downey, G., Ed.; Woodhead Publishing: Karlsruhe, Germany, 2016; pp. 565–584. [Google Scholar] [CrossRef]
- Ok, S. Detection of olive oil adulteration by low-field NMR relaxometry and UV-Vis spectroscopy upon mixing olive oil with various edible oils. Grasas Aceites 2017, 68, e173. [Google Scholar] [CrossRef] [Green Version]
- Jović, O.; Pičuljan, K.; Hrenar, T.; Smolić, T.; Primožič, I. 1H NMR adulteration study of hempseed oil with full chemometric approach on large variable data. Chemometr. Intell. Lab. 2019, 185, 41–46. [Google Scholar] [CrossRef]
- Amargianitaki, M.; Spyros, A. NMR-based metabolomics in wine quality control and authentication. Chem. Biol. Technol. Agric. 2017, 4, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Solovyev, P.A.; Fauhl-Hassek, C.; Riedl, J.; Esslinger, S.; Bontempo, L.; Camin, F. NMR spectroscopy in wine authentication: An official control perspective. Compr. Rev. Sci. Food Saf. 2021, 20, 2040–2062. [Google Scholar] [CrossRef]
- Viskić, M.; Bandić, L.M.; Korenika, A.M.J.; Jeromel, A. NMR in the service of wine differentiation. Foods 2021, 10, 120. [Google Scholar] [CrossRef]
- Lin, S.; Salcido-Keamo, S. Fraud in wine and other alcoholic beverages. In Food Fraud; Hellberg, R.S., Everstine, K., Sklare, S.A., Eds.; Academic Press: Chennai, India, 2021; pp. 233–247. [Google Scholar]
- Marchetti, L.; Pellati, F.; Benvenuti, S.; Bertelli, D. Use of 1H NMR to detect the percentage of pure fruit juices in blends. Molecules 2019, 24, 2592. [Google Scholar] [CrossRef] [Green Version]
- Chater, J.M.; Mathon, C.; Larive, C.K.; Merhaut, D.J.; Tinoco, L.W.; Mauk, P.A.; Zhenyu, J.; Preece, J.E. Juice quality traits, potassium content, and 1H NMR derived metabolites of 14 pomegranate cultivars. J. Berry Res. 2019, 9, 209–225. [Google Scholar] [CrossRef]
- Jamin, E.; Thomas, F. SNIF-NMR applications in an economic context: Fraud detection in food products. In Modern Magnetic Resonance, 2nd ed.; Webb, G.A., Ed.; Springer: New York, NY, USA, 2018; pp. 1405–1416. [Google Scholar] [CrossRef]
- Fonseca-Aguiñaga, R.; Gómez-Ruiz, H.; Miguel-Cruz, F.; Romero-Cano, L.A. Analytical characterization of tequila (silver class) using stable isotope analyses of C, O and atomic absorption as additional criteria to determine authenticity of beverage. Food Control 2020, 112, 107161. [Google Scholar] [CrossRef]
- Christodoulou, M.; Bradley, D.; Maréchal, A.; Nganga, J.; Béteille, R.; Moens, J.; Lejeune, F.; Montanari, F. Study on the mandatory indication of country of origin or place of provenance of unprocessed foods, single ingredient products and ingredients that represent more than 50% of a food. In Food Chain Evaluation Consortium for the Directorate General for Health and Food Safety; European Union: Brussels, Belgium, 2015; ISBN 978-92-79-47124-7. [Google Scholar] [CrossRef]
- Alves Filho, E.G.; Silva, L.M.A.; Ribeiro, P.R.V.; de Brito, E.S.; Zocolo, G.J.; Souza-Leão, P.C.; Marques, A.T.B.; Quintela, A.L.; Larsen, F.H.; Canuto, K.M. 1H NMR and LC-MS-based metabolomic approach for evaluation of the seasonality and viticultural practices in wines from São Francisco River Valley, a Brazilian semi-arid region. Food Chem. 2019, 289, 558–567. [Google Scholar] [CrossRef] [PubMed]
- Ün, İ.; Salim, O.K. Analysis of olive oil for authentication and shelf life determination. J. Food Sci. Techol. 2018, 55, 2476–2487. [Google Scholar] [CrossRef] [PubMed]
- Gougeon, L.; da Costa, G.; Richard, T.; Guyon, F. Wine authenticity by quantitative 1H NMR versus multitechnique analysis: A case study. Food Anal. Methods 2019, 12, 956–965. [Google Scholar] [CrossRef]
- Lia, F.; Vella, B.; Mangion, M.Z.; Farrugia, C. Application of 1H and 13C NMR fingerprinting as a tool for the authentication of Maltese extra virgin olive oil. Foods 2020, 9, 689. [Google Scholar] [CrossRef] [PubMed]
- López-Aguilar, R.; Zuleta-Prada, H.; Hernández-Montes, A.; Herbert-Pucheta, J.E. Comparative NMR metabolomics profiling between Mexican ancestral & artisanal mezcals and industrialized wines to discriminate geographical origins, agave species or grape varieties and manufacturing processes as a function of their quality attributes. Foods 2021, 10, 157. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Cao, Y.; Zhu, J.; Xu, W.; Wu, W. Analysis of metabolites in chardonnay dry white wine with various inactive yeasts by 1H NMR spectroscopy combined with pattern recognition analysis. AMB Expr. 2019, 9, 140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Forino, M.; Gambuti, A.; Moio, L. NMR-based systematic analysis of bioactive phytochemicals in red wine. First determination of xanthurenic and oleanic acids. Food Chem. 2019, 278, 497–501. [Google Scholar] [CrossRef]
- Kubicova, L.; Hadacek, F.; Bachmann, G.; Weckwerth, W.; Chobot, V. Coordination complex formation and redox properties of kynurenic and xanthurenic acid can affect brain tissue homeodynamics. Antioxidants 2019, 8, 476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, S.; Tian, X.L.; Yu, W.X.; Zhou, L.P.; Dong, X.L.; Favus, M.J.; Wong, M.S. Oleanolic acid and ursolic acid improve bone properties and calcium balance and modulate vitamin D metabolism in aged female rats. Front. Pharmacol. 2018, 9, 1435. [Google Scholar] [CrossRef]
- Ayeleso, T.B.; Matumba, M.G.; Mukwevho, E. Oleanolic acid and its derivatives: Biological activities and therapeutic potential in chronic diseases. Molecules 2017, 22, 1915. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Godelmann, R.; Fang, F.; Humpfer, E.; Schütz, B.; Bansbach, M.; Schäfer, H.; Spraul, M. Targeted and nontargeted wine analysis by 1H NMR spectroscopy combined with multivariate statistical analysis. Differentiation of important pa-rameters: Grape variety, geographical origin, year of vintage. J. Agric. Food Chem. 2013, 61, 5610–5619. [Google Scholar] [CrossRef] [PubMed]
- Monakhova, Y.; Godelmann, R.; Kuballa, T.; Mushtakova, S.; Rutledge, D. Independent components analysis to increase efficiency of discriminant analysis methods (FDA and LDA): Application to NMR fingerprinting of wine. Talanta 2015, 141, 60–65. [Google Scholar] [CrossRef] [PubMed]
- Mazzei, P.; Spaccini, R.; Francesca, N.; Moschetti, G.; Piccolo, A. Metabolomic by 1H NMR spectrosocopy differentiates “Fiano di Avellino” white wines obtained with different yeast strains. J. Agric. Food Chem. 2013, 61, 10816–10822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gougeon, L.; Da Costa, G.; Le Mao, I.; Ma, W.; Teissedre, P.; Guyon, F.; Richard, T. Wine analysis and authenticity using 1H-NMR metabolomics data: Application to Chinese wines. Food Anal. Methods 2018, 11, 3425–3434. [Google Scholar] [CrossRef]
- Hu, B.; Gao, J.; Xu, S.; Zhu, J.; Fan, X.; Zhou, X. Quality evaluation of different varieties of dry red wine based on nuclear magnetic resonance metabolomics. Appl. Biol. Chem. 2020, 63, 24. [Google Scholar] [CrossRef]
- Del Fresno, J.M.; Escott, C.; Loira, I.; Herbert-Pucheta, J.E.; Schneider, R.; Carrau, F.; Cuerda, R.; Morata, A. Impact of Hanseniaspora vineae in alcoholic fermentation and ageing on lees of high-quality white wine. Fermentation 2020, 6, 66. [Google Scholar] [CrossRef]
- Hwang, T.J.; Shaka, A.J. Water suppression that works. Excitation sculpting using arbitrary wave-forms and pulsed-field gradients. J. Magn. Reson. A 1995, 112, 275–279. [Google Scholar] [CrossRef]
- Geen, H.; Freeman, R. Band-selective radiofrequency pulses. J. Magn. Reson. 1991, 93, 93–141. [Google Scholar] [CrossRef]
- Herbert-Pucheta, J.E.; Pino-Villar, C.; Rodríguez-González, F.; Padilla-Maya, G.; Milmo-Brittingham, D.; Zepeda-Vallejo, L.G. “One-shot” analysis of wine parameters in non-Saccharomyces large-scale alcohol reduction processes with one- and two-dimensional nuclear magnetic resonance. BIO Web Conf. 2019, 15, 02016. [Google Scholar] [CrossRef]
- Herbert-Pucheta, J.E.; López-Morales, C.A.; Medina-Rivero, E.; Estrada-Parra, S.; Pérez-Tapia, S.M.; Zepeda-Vallejo, L.G. Consistency of a dialyzable leucocyte extract manufactured at GMP facilities by nuclear magnetic resonance spectroscopy. Pharm. Biomed. Anal. 2021, 196, 113940. [Google Scholar] [CrossRef] [PubMed]
- Spraul, M.; Schütz, B.; Humpfer, E.; Mörtter, M.; Schäfer, H.; Koswig, S.; Rinke, P. Mixture analysis by NMR as applied to fruit juice quality control. Magn. Reson. Chem. 2009, 47, S130–S137. [Google Scholar] [CrossRef] [PubMed]
- The AIJN Code of Practice. Available online: https://aijn.eu/en/the-aijn-code-of-practice (accessed on 30 April 2021).
- Pérez-Enciso, M.; Tenenhaus, M. Prediction of clinical outcome with microarray data: A partial least squares discriminant analysis (PLS-DA) approach. Hum. Genet. 2003, 112, 581–592. [Google Scholar] [CrossRef] [PubMed]
- Lê Cao, K.A.; Boitard, S.; Besse, P. Sparse PLS discriminant analysis: Biologically relevant feature se-lection and graphical displays for multiclass problems. BMC Bioinform. 2011, 12, 2532011. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Worley, B.; Powers, R. Multivariate analysis in metabolomics. Curr. Metabol. 2013, 1, 92–107. [Google Scholar] [CrossRef]
- Spraul, M.; Schütz, B.; Rinke, P.; Koswig, S.; Humpfer, E.; Schäfer, H.; Mörtter, M.; Fang, F.; Marx, U.C.; Minoja, A. NMR-based multi parametric quality control of fruit juices: SGF profiling. Nutrients 2009, 1, 148–155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmidtke, L.M.; Blackman, J.W. Production technologies for reduced alcoholic wines. J. Food Sci. 2012, 71, R25. [Google Scholar] [CrossRef]
- Contreras, A.; Hidalgo, C.; Schmidt, S.; Henschke, P.A.; Curtin, C.; Varela, C. The application of non-Saccharomyces yeast in fermentations with limited aeration as a strategy for the production of wine with reduced alcohol content. Int. J. Food Microbiol. 2015, 205, 7–15. [Google Scholar] [CrossRef]
- Obbi, M.; De Vero, L.; Solieri, L.; Comitini, F.; Oro, L.; Giudici, P.; Ciani, M. Fermentative aptitude of non-Saccharomyces wine yeast for reduction in the ethanol content in wine. Eur. Food Res. Technol. 2014, 239, 41–48. [Google Scholar] [CrossRef]
- Del Fresno, J.M.; Morata, A.; Loira, I.; Bañuelos, M.A.; Escott, C.; Benito, S.; Chamorro, C.G.; Suárez-Lepe, J.A. Use of non-Saccharomyces in single-culture, mixed and sequential fermentation to improve red wine quality. Eur. Food Res. Technol. 2017, 243, 2175–2185. [Google Scholar] [CrossRef]
- Benito, A.; Hofmann, T.; Laier, M.; Lochbüler, B.; Schüttler, A.; Ebert, K.; Fritsch, S.; Röcker, J.; Rauhut, D. Effect on quality and composition of Riesling wines fermented by sequential inoculation with non-Saccharomyces and Saccharomyces cerevisiae. Eur. Food Res. Technol. 2015, 241, 707–717. [Google Scholar] [CrossRef]
- Varela, C.; Barker, A.; Tran, T.; Borneman, A.; Curtin, C. Sensory profile and volatile aroma composition of reduced alcohol Merlot wines fermented with Metschnikowia pulcherrima and Saccharomyces uvarum. Int. J. Food Microbiol. 2017, 252, 1–9. [Google Scholar] [CrossRef]
- Gobbi, M.; Comitini, F.; Domizio, P.; Romani, C.; Lencioni, L.; Mannazzu, I.; Ciani, M. Lachancea thermotolerans and Saccharomyces cerevisiae in simultaneous and sequential co-fermentation: A strategy to enhance acidity and improve the overall quality of wine. Food Microbiol. 2013, 33, 271–281. [Google Scholar] [CrossRef] [PubMed]
- de Meyer, T.; Sinnaeve, D.; van Gasse, B.; Tsiporkova, E.; Rietzschel, E.; de Buyzere, M.; Gillebert, T.; Bekaert, S.; Martins, J.; van Criekinge, W. NMR-based characterization of metabolic alterations in hypertension using an adaptive, intelligent binning algorithm. Anal. Chem. 2008, 80, 3783–3790. [Google Scholar] [CrossRef] [PubMed]
- Shishmarev, D.; Otting, G. Radiation damping on cryoprobes. J. Magn. Reason. 2011, 213, 76–81. [Google Scholar] [CrossRef] [PubMed]
- Ebrahimi, P.; Viereck, N.; Bro, R.; Engelsen, S.B. Chemometric Analysis of NMR Spectra. In Modern Magnetic Resonance, 2nd ed.; Webb, G.A., Ed.; Springer: Cham, Switzerland, 2018; pp. 1–20. [Google Scholar] [CrossRef]
- Jacob, D.; Deborde, C.; Lefevbre, M.; Maucourt, M.; Moing, A. NMRProcFlow: A graphical and interactive tool dedicated to 1D spectra processing for NMR-based metabolomics. Metabolomics 2017, 13, 36. [Google Scholar] [CrossRef] [Green Version]
- Chong, J.; Wishart, D.S.; Xia, J. Using MetaboAnalyst 4.0 for comprehensive and integrative metabolomics data analysis. Curr. Protoc. Bioinform. 2019, 68, e86. [Google Scholar] [CrossRef]
- Holmes, E.; Loo, R.L.; Stamler, J.; Bictash, M.; Yap, I.K.S.; Chan, Q.; Ebbels, T.; De Iorio, M.; Brown, I.J.; Veselkov, K.A.; et al. Human metabolic phenotype diversity and its association with diet and blood pressure. Nature 2008, 453, 396–400. [Google Scholar] [CrossRef]
- Xu, Q.-S.; Liang, Y.-Z. Monte Carlo cross validation. Chemom. Intell. Lab. Syst. 2001, 56, 1–11. [Google Scholar] [CrossRef]








| Compound | Commercial Apple Juice (Not from Concentrate) | Apple Juice Made from Concentrate | Orange Juice Made from Concentrate | Apricot Juice Made from Concentrate | ||||
|---|---|---|---|---|---|---|---|---|
| Conc. | Unit | Conc. | Unit | Conc. | Unit | Conc. | Unit | |
| Ethanol | 195 | mg/L | <10 | mg/L | <10 | mg/L | 11 | mg/Kg |
| Lactic acid | 66 | mg/L | <10 | mg/L | 77 | mg/L | <20 | mg/Kg |
| 5-Hydroxymethylfurfural | 6 | mg/L | <5 | mg/L | <5 | mg/L | <5 | mg/Kg |
| Citric acid | <0.5 | g/L | <0.5 | g/L | 6.7 | g/L | 7.2 | g/Kg |
| Malic acid | 4.3 | g/L | 1.6 | g/L ↓ | 1.5 | g/L | 4.9 | g/Kg ↓ |
| Glucose | 21.5 | g/L | 8.6 | g/L ↓ | 18.6 | g/L ↓ | 32.2 | g/Kg |
| Fructose | 62.9 | g/L | 21.8 | g/L ↓ | 20.9 | g/L | 19.5 | g/Kg |
| Glucose/fructose ratio | 0.34 | 0.39 | 0.89 | 1.65 | ||||
| Sucrose | 14.1 | g/L | 7 | g/L | 50.1 | g/L ↑ | 16 | g/Kg |
| % Sucrose | 14 | % | 19 | % | 56 | % ↑ | 24 | % |
| Total sugar | 98.6 | g/L | 37.3 | g/L | 89.6 | g/L | 67.7 | g/Kg |
| Alanine | 20 | mg/L | <5 | mg/L ↓ | 39 | mg/L ↓ | 28 | mg/Kg ↓ |
| Acetaldehyde | <5 | mg/L | <5 | mg/L | <5 | mg/L | <5 | mg/Kg |
| Benzoic acid | <10 | mg/L | <10 | mg/L | <10 | mg/L | <10 | mg/Kg |
| Formic acid | <5 | mg/L | <5 | mg/L | <5 | mg/L | 5 | mg/Kg |
| Methanol | 44 | mg/L | <10 | mg/L | <10 | mg/L | 40 | mg/Kg |
| Sorbic acid | <10 | mg/L | <10 | mg/L | <10 | mg/L | <10 | mg/Kg |
| Succinic acid | 16 | mg/L | <10 | mg/L | 17 | mg/L | 32 | mg/Kg |
| Benzaldehyde | <5 | mg/L | <5 | mg/L | <5 | mg/L | ND | |
| Proline | <50 | mg/L | <50 | mg/L | 366 | mg/L ↓ | ND | |
| Galacturonic acid | 648 | mg/L | <100 | mg/L | <150 | mg/L | ND | |
| Acetone | <10 | mg/L | <10 | mg/L | ND | ND | ||
| Arbutin | <10 | mg/L | <10 | mg/L | ND | ND | ||
| Chlorogenic acid | 28 | mg/L | 53 | mg/L | ND | ND | ||
| Citramalic acid | 33 | mg/L | <10 | mg/L | ND | ND | ||
| Malic/Quinic ratio | 17.4 | mg/L | 20.5 | mg/L | ND | ND | ||
| Fumaric acid | <5 | mg/L | <5 | mg/L | ND | ND | ||
| Pyruvic acid | <10 | mg/L | <10 | mg/L | ND | ND | ||
| Quinic acid | 247 | mg/L | 76 | mg/L | ND | ND | ||
| Xylose | 529 | mg/L | <300 | mg/L | ND | ND | ||
| Potassium * | 1016 | mg/L | ND | ND | ND | |||
| Magnesium * | 39 | mg/L ↓ | ND | ND | ND | |||
| Titratable Acidity pH 7 * | 51 | meq/L | ND | ND | ND | |||
| Titratable Acidity pH 8.1 * | 52 | meq/L | ND | ND | ND | |||
| Titratable Acidity (pH 7, tartaric acid) * | 3.8 | g/L | ND | ND | ND | |||
| Titratable Acidity (pH 7, malic acid) * | 3.3 | g/L | ND | ND | ND | |||
| Titratable Acidity (pH 8.1, citric acid) * | 3.3 | g/L | ND | ND | ND | |||
| 4-Aminobutanoic acid | ND | ND | 163 | mg/L ↓ | ND | |||
| Arginine | ND | ND | 622 | mg/L | ND | |||
| Phlorin | ND | ND | <10 | mg/L | ND | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Herbert-Pucheta, J.E.; Lozada-Ramírez, J.D.; Ortega-Regules, A.E.; Hernández, L.R.; Anaya de Parrodi, C. Nuclear Magnetic Resonance Metabolomics with Double Pulsed-Field-Gradient Echo and Automatized Solvent Suppression Spectroscopy for Multivariate Data Matrix Applied in Novel Wine and Juice Discriminant Analysis. Molecules 2021, 26, 4146. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules26144146
Herbert-Pucheta JE, Lozada-Ramírez JD, Ortega-Regules AE, Hernández LR, Anaya de Parrodi C. Nuclear Magnetic Resonance Metabolomics with Double Pulsed-Field-Gradient Echo and Automatized Solvent Suppression Spectroscopy for Multivariate Data Matrix Applied in Novel Wine and Juice Discriminant Analysis. Molecules. 2021; 26(14):4146. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules26144146
Chicago/Turabian StyleHerbert-Pucheta, José Enrique, José Daniel Lozada-Ramírez, Ana E. Ortega-Regules, Luis Ricardo Hernández, and Cecilia Anaya de Parrodi. 2021. "Nuclear Magnetic Resonance Metabolomics with Double Pulsed-Field-Gradient Echo and Automatized Solvent Suppression Spectroscopy for Multivariate Data Matrix Applied in Novel Wine and Juice Discriminant Analysis" Molecules 26, no. 14: 4146. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules26144146