Synthesis, Structure and Impact of 5-Aminoorotic Acid and Its Complexes with Lanthanum(III) and Gallium(III) on the Activity of Xanthine Oxidase
Abstract
:1. Introduction
2. Results
2.1. Chemistry
2.1.1. Vibrational Spectroscopy
2.1.2. 1H NMR Spectra of the Ligand and Its Metal Complexes
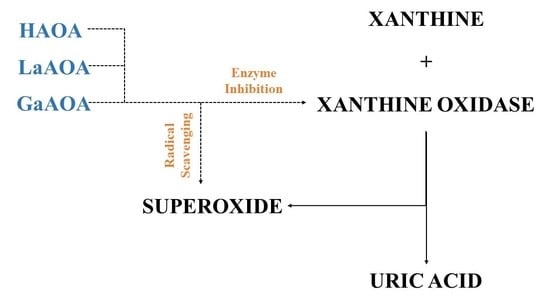
2.2. Radical-Scavenging Assays
2.3. Molecular Docking Assay
3. Discussion and Conclusions
- HAOA, LaAOA, and GaAOA are all scavengers of superoxide, derived from KO2 and X/XO;
- All three substances manifest higher superoxide-diminishing activity in the non-enzymatic model, compared to the enzymatic model;
- All three substances diminish the production of UA in the X/XO model. Previously, we proposed that this may be due to an interaction between the substances and one/both elements of the model system;
- Molecular docking suggested that all three substances interact with the active sites of XO, showing potential as enzyme inhibitors. The calculated strength of the interaction decreases in the order LaAOA > GaAOA > HAOA. This result corresponds well with the data from the XO activity UV assay—HAOA decreases UA formation by 15–20%, while both complexes cause a decrease of about 40–45%.
- In the enzymatic models, superoxide diminishment is greater than the suppression of UA production. Since, for one molecule UA produced, one superoxide ion is generated, we can propose with a reasonable amount of certainty that in the X/XO model, HAOA, GaAOA, and LaAOA act as both:
- superoxide scavengers, which is clear enough from the results, derived from the KO2 assay;
- XO inhibitors, a proposition supported by the decrease in UA production in the X/XO model and also by the molecular docking assay. Another possible confirmation of that hypothesis comes from the fact that as molecular docking identified HAOA as the substance with weakest interaction with XO, that same substance caused the least diminishing of UA production in the enzymatic model.
4. Materials and Methods
4.1. Synthesis of the Complexes
4.2. Analytical and Spectroscopic Methods
4.3. Radical-Scavenging Assays
4.3.1. Assay for CL in the Presence of KO2
4.3.2. Assay for CL in the Presence of X/XO Model System
4.3.3. Assay for UV Determination of XO Activity in the Presence of the X/XO Model System
4.4. Statistical Analysis
In Silico Molecular Docking Assay
- removing ligands, heteroatoms, and unnecessary water molecules unless needed at the active site (step I);
- checking the receptor ionization and tautomeric states by adding the necessary hydrogen atoms (step II);
- defining the active site by using the list of active site residues mode (step III);
- setting the ligands by performing energy minimization using the Hyperchem v8.0 program [100]. First, we used the single point mode to calculate the ligand energy and gradient, then molecular mechanics optimization was performed using the Polack–Ribiere conjugate gradient algorithm where the RMS gradient of 0.1 kcal/(Å mol) was used for 735 cycles (step IV);
- fitness function choice, and the piecewise linear potential (PLPChem score) was chosen as the most recommended fitness function in GOLD [99] (step V);
- the number of genetic algorithm (GA) runs, and 100 solutions (results) per inhibitor were chosen as the most accurate (step VI).
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Arouma, O. Free radicals, oxidative stress, and antioxidants in human health and disease. Nutrition 2002, 18, 872–879. [Google Scholar] [CrossRef]
- Valko, M.; Leibfritz, D.; Moncol, J.; Cronin, M.T.; Mazur, M.; Telser, J. Free radicals and antioxidants in normal physiological functions and human disease. Int. J. Biochem. Cell Biol. 2007, 39, 44–84. [Google Scholar] [CrossRef] [PubMed]
- Guzik, T.J.; Korbut, R.; Adamek-Guzik, T. Nitric oxide and superoxide in inflammation and immune regulation. J. Physiol. Pharmacol. 2003, 54, 469–487. [Google Scholar]
- Ullrich, V.; Namgaladze, D.; Frein, D. Superoxide as inhibitor of calcineurin and mediator of redox regulation. Toxicol. Lett. 2003, 139, 107–110. [Google Scholar] [CrossRef]
- Linnane, A.W.; Kios, M.; Vitetta, L. The essential requirement for superoxide radical and nitric oxide formation for normal physiological function and healthy aging. Mitochondrion 2007, 7, 1–5. [Google Scholar] [CrossRef]
- Ullrich, V.; Bachschmid, M. Superoxide as a messenger of endothelial function. Biochem. Biophys. Res. Commun. 2000, 278, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mayorov, D.N. Brain superoxide as a key regulator of the cardiovascular response to emotional stress in rabbits. Exp. Physiol. 2007, 92, 471–479. [Google Scholar] [CrossRef]
- Brand, M.D. Mitochondrial generation of superoxide and hydrogen peroxide as the source of mitochondrial redox signaling. Free Radic. Biol. Med. 2016, 100, 14–31. [Google Scholar] [CrossRef]
- Porporato, P.E.; Payen, V.L.; Pérez-Escuredo, J.; De Saedeleer, C.J.; Danhier, P.; Copetti, T.; Dhup, S.; Tardy, M.; Vazeille, T.; Bouzin, C.; et al. A mitochondrial switch promotes tumor metastasis. Cell Rep. 2014, 8, 754–766. [Google Scholar] [CrossRef] [Green Version]
- Kosmider, B.; Lin, C.-R.; Karim, L.; Tomar, D.; Vlasenko, L.; Marchetti, N.; Bolla, S.; Madesh, M.; Criner, G.J.; Bahmed, K. Mitochondrial dysfunction in human primary alveolar type II cells in emphysema. EBioMedicine 2019, 46, 305–316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, T.; Fan, S.; Zheng, D.; Wang, G.; Yu, Y.; Chen, R.; Song, L.-S.; Fan, G.-C.; Zhang, Z.; Peng, T. Increased calpain-1 in mitochondria induces dilated heart failure in mice: Role of mitochondrial superoxide anion. Basic Res. Cardiol. 2019, 114, 1–15. [Google Scholar] [CrossRef]
- Suski, J.M.; Lebiedzinska, M.; Bonora, M.; Pinton, P.; Duszynski, J.; Wieckowski, M.R. Relation between mitochondrial membrane potential and ROS formation. In Mitochondrial Bioenergetics; Springer: Berlin/Heidelberg, Germany, 2012; pp. 183–205. [Google Scholar]
- Kalyanaraman, B.; Cheng, G.; Hardy, M.; Ouari, O.; Bennett, B.; Zielonka, J. Teaching the basics of reactive oxygen species and their relevance to cancer biology: Mitochondrial reactive oxygen species detection, redox signaling, and targeted therapies. Redox Biol. 2018, 15, 347–362. [Google Scholar] [CrossRef] [PubMed]
- Matés, J.M.; Segura, J.A.; Alonso, F.J.; Márquez, J. Oxidative stress in apoptosis and cancer: An update. Arch. Toxicol. 2012, 86, 1649–1665. [Google Scholar] [CrossRef] [PubMed]
- Indo, H.P.; Yen, H.-C.; Nakanishi, I.; Matsumoto, K.-I.; Tamura, M.; Nagano, Y.; Matsui, H.; Gusev, O.; Cornette, R.; Okuda, T.; et al. A mitochondrial superoxide theory for oxidative stress diseases and aging. J. Clin. Biochem. Nutr. 2015, 56, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Hayyan, M.; Hashim, M.A.; AlNashef, I.M. Superoxide ion: Generation and chemical implications. Chem. Rev. 2016, 116, 3029–3085. [Google Scholar] [CrossRef] [Green Version]
- Murrell, G.A.; Francis, M.J.; Bromley, L. Modulation of fibroblast proliferation by oxygen free radicals. Biochem. J. 1990, 265, 659–665. [Google Scholar] [CrossRef] [Green Version]
- Weening, R.; Wever, R.; Roos, D. Quantitative aspects of the production of superoxide radicals by phagocytizing human granulocytes. J. Lab. Clin. Med. 1975, 85, 245–252. [Google Scholar] [PubMed]
- Matsubara, T.; Ziff, M. Increased superoxide anion release from human endothelial cells in response to cytokines. J. Immunol. 1986, 137, 3295–3298. [Google Scholar]
- Burdon, R.H. Superoxide and hydrogen peroxide in relation to mammalian cell proliferation. Free Rad. Biol. Med. 1995, 18, 775–794. [Google Scholar] [CrossRef]
- Day, R.M.; Suzuki, Y.J. Cell proliferation, reactive oxygen and cellular glutathione. Dose-Response 2005, 3. [Google Scholar] [CrossRef]
- Valko, M.; Rhodes, C.; Moncol, J.; Izakovic, M.; Mazur, M. Free radicals, metals and antioxidants in oxidative stress-induced cancer. Chem. Biol. Interact. 2006, 160, 1–40. [Google Scholar] [CrossRef]
- Prasad, S.; Gupta, S.C.; Tyagi, A.K. Reactive oxygen species (ROS) and cancer: Role of antioxidative nutraceuticals. Cancer Lett. 2017, 387, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Dayem, A.A.; Choi, H.-Y.; Kim, J.-H.; Cho, S.-G. Role of oxidative stress in stem, cancer, and cancer stem cells. Cancers 2010, 2, 859–884. [Google Scholar] [CrossRef] [Green Version]
- Tafani, M.; Sansone, L.; Limana, F.; Arcangeli, T.; De Santis, E.; Polese, M.; Fini, M.; Russo, M.A. The interplay of reactive oxygen species, hypoxia, inflammation, and sirtuins in cancer initiation and progression. Oxid. Med. Cell. Longev. 2016, 2016, 1–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nazarewicz, R.R.; Dikalova, A.; Bikineyeva, A.; Ivanov, S.; Kirilyuk, I.A.; Grigor’ev, I.A.; Dikalov, S.I. Does Scavenging of Mitochondrial Superoxide Attenuate Cancer Prosurvival Signaling Pathways? Mary Ann Liebert, Inc.: New Rochelle, NY, USA, 2013. [Google Scholar]
- Shah, M.H.; Liu, G.-S.; Thompson, E.W.; Dusting, G.J.; Peshavariya, H.M. Differential effects of superoxide dismutase and superoxide dismutase/catalase mimetics on human breast cancer cells. Breast Cancer Res. Treat. 2015, 150, 523–534. [Google Scholar] [CrossRef] [PubMed]
- Gill, J.G.; Piskounova, E.; Morrison, S.J. (Eds.) Cancer, Oxidative Stress, and Metastasis. Cold Spring Harbor Symposia on Quantitative Biology; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2016. [Google Scholar]
- Sosa, V.; Moliné, T.; Somoza, R.; Paciucci, R.; Kondoh, H.; LLeonart, M.E. Oxidative stress and cancer: An overview. Ageing Res. Rev. 2013, 12, 376–390. [Google Scholar] [CrossRef] [PubMed]
- Luis, A.; Sandalio, L.M.; Palma, J.; Bueno, P.; Corpas, F.J. Metabolism of oxygen radicals in peroxisomes and cellular implications. Free Radic. Biol. Med. 1992, 13, 557–580. [Google Scholar]
- Kostić, D.A.; Dimitrijević, D.S.; Stojanović, G.S.; Palić, I.R.; Đorđević, A.S.; Ickovski, J.D. Xanthine oxidase: Isolation, assays of activity, and inhibition. J. Chem. 2015, 2015. [Google Scholar] [CrossRef] [Green Version]
- Cos, P.; Ying, L.; Calomme, M.; Hu, J.P.; Cimanga, K.; Van Poel, B.; Pieters, L.; Vlietinck, A.J.; Berghe, D.V. Structure−activity relationship and classification of flavonoids as inhibitors of xanthine oxidase and superoxide scavengers. J. Nat. Prod. 1998, 61, 71–76. [Google Scholar] [CrossRef]
- Mittal, A.; Phillips, A.R.; Loveday, B.; Windsor, J.A. The potential role for xanthine oxidase inhibition in major intra-abdominal surgery. World J. Surg. 2008, 32, 288–295. [Google Scholar] [CrossRef]
- Candan, F. Effect of Rhus coriaria L.(Anacardiaceae) on superoxide radical scavenging and xanthine oxidase activity. J. Enzyme Inhib. Med. Chem. 2003, 18, 59–62. [Google Scholar] [CrossRef]
- Kelley, E.E.; Khoo, N.K.; Hundley, N.J.; Malik, U.Z.; Freeman, B.A.; Tarpey, M.M. Hydrogen peroxide is the major oxidant product of xanthine oxidase. Free Rad. Biol. Med. 2010, 48, 493–498. [Google Scholar] [CrossRef] [Green Version]
- Xu, H.; Li, C.; Mozziconacci, O.; Zhu, R.; Xu, Y.; Tang, Y.; Chen, R.; Huang, Y.; Holzbeierlein, J.M.; Schöneich, C.; et al. Xanthine oxidase-mediated oxidative stress promotes cancer cell-specific apoptosis. Free Rad. Biol. Med. 2019, 139, 70–79. [Google Scholar] [CrossRef]
- Oh, S.-H.; Choi, S.-Y.; Choi, H.-J.; Ryu, H.-M.; Kim, Y.-J.; Jung, H.-Y.; Cho, J.-H.; Kim, C.-D.; Park, S.-H.; Kwon, T.-H.; et al. The emerging role of xanthine oxidase inhibition for suppression of breast cancer cell migration and metastasis associated with hypercholesterolemia. FASEB J. 2019, 33, 7301–7314. [Google Scholar] [CrossRef]
- Ogura, J.; Kuwayama, K.; Sasaki, S.; Kaneko, C.; Koizumi, T.; Yabe, K.; Tsujimoto, T.; Takeno, R.; Takaya, A.; Kobayashi, M.; et al. Reactive oxygen species derived from xanthine oxidase interrupt dimerization of breast cancer resistance protein, resulting in suppression of uric acid excretion to the intestinal lumen. Biochem. Pharmacol. 2015, 97, 89–98. [Google Scholar] [CrossRef] [PubMed]
- Furue, H. Chemotherapy cancer treatment during the past sixty years. Cancer Chemother. 2003, 30, 1404–1411. [Google Scholar]
- Wiemann, M.; Calabresi, P. Principles of current cancer chemotherapy. Compr. Ther. 1983, 9, 46–52. [Google Scholar] [PubMed]
- Tanneberger, S. Cancer chemotherapy today and in the future. Z. Gesamte Inn. Med. Grenzgeb. 1990, 45, 693–695. [Google Scholar]
- Kim, S.J.; Kim, H.S.; Seo, Y.R. Understanding of ROS-inducing strategy in anticancer therapy. Oxid. Med. Cell. Longev. 2019, 2019, 5381692. [Google Scholar] [CrossRef]
- Wang, K.; Li, R.; Cheng, Y.; Zhu, B. Lanthanides—The future drugs? Coord. Chem. Rev. 1999, 190, 297–308. [Google Scholar] [CrossRef]
- Kostova, I. Lanthanides as anticancer agents. Curr. Med. Chem. 2005, 5, 591–602. [Google Scholar] [CrossRef]
- Todorov, L.; Kostova, I.; Traykova, M. Lanthanum, Gallium and their Impact on Oxidative Stress. Curr. Med. Chem. 2019, 26, 4280–4295. [Google Scholar] [CrossRef]
- Lessa, J.A.; Parrilha, G.L.; Beraldo, H. Gallium complexes as new promising metallodrug candidates. Inorg. Chim. Acta 2012, 393, 53–63. [Google Scholar] [CrossRef]
- Kapoor, S. Lanthanum and its rapidly emerging role as an anti-carcinogenic agent. J. Cell. Biochem. 2009, 106, 193. [Google Scholar] [CrossRef] [PubMed]
- Durgo, K.; Halec, I.; Šola, I.; Franekić, J. Cytotoxic and genotoxic effects of the quercetin/lanthanum complex on human cervical carcinoma cells in vitro. Arhiv za Higijenu Rada i Toksikologiju 2011, 62, 221–226. [Google Scholar] [CrossRef] [Green Version]
- Shen, L.; Lan, Z.; Sun, X.; Shi, L.; Liu, Q.; Ni, J. Proteomic analysis of lanthanum citrate-induced apoptosis in human cervical carcinoma SiHa cells. Biometals 2010, 23, 1179–1189. [Google Scholar] [CrossRef]
- Valcheva-Traykova, M.; Saso, L.; Kostova, I. Involvement of lanthanides in the free radicals’ homeostasis. Curr. Top. Med. Chem. 2014, 14, 2508–2519. [Google Scholar] [CrossRef] [PubMed]
- Todorov, L.; Chifchiev, B.; Valcheva-Traykova, M.; Kostova, I. Radical scavenging activity toward 2, 2-diphenyl-1-picrylhydrazyl and hydroxyl radicals of 5 aminoorotic acid and its Ga (III) complex. Bulg. Chem. Commun. 2018, 50, 207–212. [Google Scholar]
- Todorov, L.; Valcheva-Traykova, M.; Traykov, T.; Kostova, I. Impact of 5-aminoorotic acid and its complex with gallium (III) on the luminol-dependent chemiluminescence in presence of sodium hypochlorite. AIP Conf. Proc. 2019, 2075, 170004. [Google Scholar]
- Todorov, L.; Valcheva-Traykova, M.; Atanasova, V.; Kostova, I. Effect of 5-aminoorotic acid and its gallium (III) complex on the antioxidant activity of rat blood serum. Bulg. Chem. Commun. 2019, 51, 200–203. [Google Scholar]
- Todorov, L.; Valcheva-Traykova, M.; Kostova, I. In Vitro Interaction of 5-aminoorotic Acid and Its Lantnanum(III) Complex With Superoxide and Hypochlorite Radicals. Pharma Chem. 2020, 12, 10. [Google Scholar]
- Todorov, L.; Traykova, M.; Saso, L.; Kostova, I. In Vitro Interaction of 5-Aminoorotic Acid and Its Gallium (III) Complex with Superoxide Radical, Generated by Two Model Systems. Int. J. Mol. Sci. 2020, 21, 8862. [Google Scholar] [CrossRef]
- Hernanz, A.; Billes, F.; Bratu, I.; Navarro, R. Vibrational analysis and spectra of orotic acid. Biopolymers 2000, 57, 187–198. [Google Scholar] [CrossRef]
- Takusagawa, F.; Shimada, A. The crystal structure of orotic acid monohydrate (Vitamin B13). Bull. Chem. Soc. Jpn. 1973, 46, 2011–2019. [Google Scholar] [CrossRef] [Green Version]
- Hilal, R.; Zaky, Z.; Elroby, S.A. Electronic structure of orotic acid I. Geometry, conformational preference and tautomerism. J. Mol. Struct. THEOCHEM 2004, 685, 35–42. [Google Scholar]
- Dinda, J.; Bag, K.; Sinha, C.; Mostafa, G.; Lu, T.-H. Naphthylazoimidazole and mercury (II) complexes. Single crystal X-ray structure of 1-ethyl-2-(naphthyl-α-azo) imidazolium hexaflurophosphate. Polyhedron 2003, 22, 1367–1376. [Google Scholar] [CrossRef]
- Wysokiński, R.; Morzyk-Ociepa, B.; Głowiak, T.; Michalska, D. Revised molecular structure and vibrational spectra of tetraaqua (orotato) nickel (II) monohydrate: Band assignment based on density functional calculations. J. Mol. Struct. 2002, 606, 241–251. [Google Scholar] [CrossRef]
- Allen, F.H.; Kennard, O.; Watson, D.G.; Brammer, L.; Orpen, A.G.; Taylor, R. Tables of bond lengths determined by X-ray and neutron diffraction. Part 1. Bond lengths in organic compounds. J. Chem. Soc. Perkin Trans. 2 1987, S1–S19. [Google Scholar] [CrossRef]
- Baran, E.J.; Mercader, R.C.; Hueso-Uren, F.; Moreno-Carretero, M.N.; Quiros-Olozabal, M.; Salas-Peregrin, J.M. Crystal structure, raman and 57Fe mo¨ ssbauer spectra of the FeII complex of iso-orotic acid. Polyhedron 1996, 15, 1717–1721. [Google Scholar] [CrossRef]
- Schneider, A.G.; Schmalle, H.W.; Arod, F.; Dubler, E. The interaction of 5-fluoroorotic acid with transition metals: Synthesis and characterisation of Ni (II), Cu (II) and Zn (II) complexes. J. Inorg. Biochem. 2002, 89, 227–236. [Google Scholar] [CrossRef]
- Allen, F.H.; Kirby, A.J. Bond length and reactivity. Variable length of the carbon-oxygen single bond. J. Am. Chem. Soc. 1984, 106, 6197–6200. [Google Scholar] [CrossRef]
- Suter, H.U.; Nonella, M. A Quantum Chemical Investigation of the C−O Bond Length and Stretching Mode of the Phenolate Anion. J. Phys. Chem. A 1998, 102, 10128–10133. [Google Scholar] [CrossRef]
- Djordjevic, C.; Vuletic, N.; Jacobs, B.A.; Lee-Renslo, M.; Sinn, E. Molybdenum (VI) peroxo α-amino acid complexes: Synthesis, spectra, and properties of MoO(O2)2 (α-aa) (H2O) for α-aa = glycine, alanine, proline, valine, leucine, serine, asparagine, glutamine, and glutamic acid. X-ray crystal structures of the glycine, alanine, and proline compounds. Inorg. Chem. 1997, 36, 1798–1805. [Google Scholar]
- Kieninger, M.; Ventura, O.N.; Suhai, S. Density functional investigations of carboxyl free radicals: Formyloxyl, acetyloxyl, and benzoyloxyl radicals. Int. J. Quantum Chem. 1998, 70, 253–267. [Google Scholar] [CrossRef]
- Darensbourg, D.J.; Draper, J.D.; Larkins, D.L.; Frost, B.J.; Reibenspies, J.H. Organometallic Derivatives of Orotic Acid. CO− Labilizing Ability of the Amido Group in Chromium and Tungsten Carbonyl Complexes. Inorg. Chem. 1998, 37, 2538–2546. [Google Scholar] [CrossRef]
- Bekiroglu, S.; Kristiansson, O. Hydrogen-bonded neutral and anionic lamellar networks: Crystal structures of bis (O, O′, O″-hydroorotato) disilver (i) dihydrate, potassium hydroorotate and rubidium hydroorotate. Ab initio calculations on orotic acid and the hydroorotate anion. J. Chem. Soc. Dalton Trans. 2002, 1330–1335. [Google Scholar] [CrossRef]
- Arrizabalaga, P.; Castan, P.; Dahan, F. Coordination sites of 5-nitro-6-carboxyuracil: UV study and x-ray structure determination of diammine (5-nitroorotato) copper (II) hydrate and hexaamminebis (5-nitroorotato) tricopper (II) pentahydrate. Inorg. Chem. 1983, 22, 2245–2252. [Google Scholar] [CrossRef]
- Lewandowski, W.; Kalinowska, M.; Lewandowska, H. The influence of metals on the electronic system of biologically important ligands. Spectroscopic study of benzoates, salicylates, nicotinates and isoorotates. Review. J. Inorg. Biochem. 2005, 99, 1407–1423. [Google Scholar] [CrossRef]
- Lalioti, N.; Raptopoulou, C.P.; Terzis, A.; Panagiotopoulos, A.; Perlepes, S.P.; Manessi-Zoupa, E. New metal-binding modes for 5-aminoorotic acid: Preparation, characterization and crystal structures of zinc (II) complexes. J. Chem. Soc. Dalton Trans. 1998, 1327–1334. [Google Scholar] [CrossRef]
- Dega-Szafran, Z.; Dulewicz, E.; Dutkiewicz, G.; Kosturkiewicz, Z.; Szafran, M. Two polymorphs of 4-hydroxy-1-methylpiperidine betaine hydrochloride studied by X-ray and DFT methods. J. Mol. Struct. 2005, 751, 139–150. [Google Scholar] [CrossRef]
- Horn, K.H.; Böres, N.; Lehnert, N.; Mersmann, K.; Näther, C.; Peters, G.; Tuczek, F. Reduction Pathway of End-On Terminally Coordinated Dinitrogen. IV. Geometric, Electronic, and Vibrational Structure of a W (IV) Dialkylhydrazido Complex and Its Two-Electron-Reduced Derivative Undergoing N−N Cleavage upon Protonation. Inorg. Chem. 2005, 44, 3016–3030. [Google Scholar] [CrossRef]
- Batt, R.D.; Martin, J.K.; Ploeser, J.M.; Murray, J. Chemistry of the Dihydropyrimidines. Ultraviolet Spectra and Alkaline Decomposition1a. J. Am. Chem. Soc. 1954, 76, 3663–3665. [Google Scholar] [CrossRef]
- Icbudak, H.; Olmez, H.; Yesilel, O.Z.; Arslan, F.; Naumov, P.; Jovanovski, G.; Ibrahim, A.R.; Usman, A.; Fun, H.K.; Chantrapromma, S.; et al. Syntheses, characterization and crystal structures of novel amine adducts of metal saccharinates, orotates and salicylates. J. Mol. Struct. 2003, 657, 255–270. [Google Scholar] [CrossRef]
- Papaefstathiou, G.S.; Manessi, S.; Raptopoulou, C.P.; Behrman, E.J.; Zafiropoulos, T.F. The first metal complex of 5-hydroxyorotic acid: Dimethylammonium bis (N, N-dimethylformamide) bis (5-hydroxyorotato (-2)) gallate (III). Inorg. Chem. Commun. 2004, 7, 69–72. [Google Scholar] [CrossRef]
- Lacher, J.; Bitner, J.; Park, J. The Infrared Absorption Spectra of Some Antibiotics in Antimony Trichloride Solution. J. Phys. Chem. 1955, 59, 610–614. [Google Scholar] [CrossRef]
- Lencioni, S.; Pellerito, A.; Fiore, T.; Giuliani, A.; Pellerito, L.; Cambria, M.T.; Mansueto, C. Organometallic complexes with biological molecules. X: Dialkyltin (IV) and trialkyltin (IV) orotates: Spectroscopic and in vivo investigations. Appl. Organomet. Chem. 1999, 13, 145–157. [Google Scholar] [CrossRef]
- Kostova, I.; Peica, N.; Kiefer, W. Theoretical and spectroscopic studies of lanthanum (III) complex of 5-aminoorotic acid. Chem. Phys. 2006, 327, 494–505. [Google Scholar] [CrossRef]
- Lewandowski, W.; Barańska, H. Comparison of the influence of silver, iron (III) and chromium (III) on the aromatic system of benzoic and salicylic acids in hydrated and anhydrous complexes. Vib. Spectrosc. 1991, 2, 211–220. [Google Scholar] [CrossRef]
- Lewandowski, W.; Dasiewicz, B.; Koczoń, P.; Skierski, J.; Dobrosz-Teperek, K.; Świsłocka, R.; Fuks, L.; Priebe, W.; Mazurek, A. Vibrational study of alkaline metal nicotinates, benzoates and salicylates. J. Mol. Struct. 2002, 604, 189–193. [Google Scholar] [CrossRef]
- Koczon, P.; Lewandowski, W.; Mazurek, A. Vibrational (FT-IR and FT-Raman) and NMR studies on selected metal (Ca, Mn, Zn) complexes with ortho-, meta-, and para-iodobenzoic acids. Vib. Spectrosc. 1999, 20, 143–149. [Google Scholar] [CrossRef]
- Kakiuchi, M.; Abe, T.; Nakayama, H. D/H fractionation factor between water vapor and crystal water of copper chloride dihydrate: Statistical mechanical approach based on Raman spectra. Geochem. J. 2001, 35, 285–293. [Google Scholar] [CrossRef] [Green Version]
- Wang, K.; Li, Y.-S. Silver doping of polycarbonate films for surface-enhanced Raman scattering. Vib. Spectrosc. 1997, 14, 183–188. [Google Scholar] [CrossRef]
- Boerio, F.; Hong, P.; Clark, P.; Okamoto, Y. Surface-enhanced Raman scattering from model acrylic adhesive systems. Langmuir 1990, 6, 721–727. [Google Scholar] [CrossRef]
- Kwon, Y.J.; Son, D.H.; Ahn, S.J.; Kim, M.S.; Kim, K. Vibrational spectroscopic investigation of benzoic acid adsorbed on silver. J. Phys. Chem. 1994, 98, 8481–8487. [Google Scholar] [CrossRef]
- Gałdecka, E.; Gałdecki, Z.; Huskowska, E.; Amirkhanov, V.; Legendziewicz, J. Crystal structure and optical properties of Ln (III) octahedral complexes with hexamethylphosphortriamide; [Ln (HMPA)6](ClO4)3. J. Alloys Compd. 1997, 257, 182–190. [Google Scholar] [CrossRef]
- De Andres, A.; Taboada, S.; Martínez, J.; Salinas, A.; Hernández, J.; Sáez-Puche, R. Optical phonons in R2BaMO5 oxides with M = Co, Ni, Cu, and R = a rare earth. Phys. Rev. B 1993, 47, 14898. [Google Scholar] [CrossRef]
- Cho, B.-O.; Lao, S.X.; Chang, J.P. Origin and effect of impurity incorporation in plasma-enhanced ZrO2 deposition. J. Appl. Phys. 2003, 93, 9345–9351. [Google Scholar] [CrossRef]
- Barfield, M. Proton and Carbon-13 nmr Spectroscopy; Abraham, R.J., Loftus, P., Eds.; ACS Publications: Washington, DC, USA, 1979. [Google Scholar]
- Jackman, L.M.; Sternhell, S. Application of Nuclear Magnetic Resonance Spectroscopy in Organic Chemistry: International Series in Organic Chemistry; Elsevier: Amsterdam, The Netherlands, 2013. [Google Scholar]
- Enroth, C.; Eger, B.T.; Okamoto, K.; Nishino, T.; Nishino, T.; Pai, E.F. Crystal structures of bovine milk xanthine dehydrogenase and xanthine oxidase: Structure-based mechanism of conversion. Proc. Natl. Acad. Sci. USA 2000, 97, 10723–10728. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Linani, A.; Benarous, K.; Bou-Salah, L.; Yousfi, M. Hispidin, Harmaline, and Harmine as potent inhibitors of bovine xanthine oxidase: Gout treatment, in vitro, ADMET prediction, and SAR studies. Bioorg. Chem. 2021, 112, 104937. [Google Scholar] [CrossRef] [PubMed]
- Bou-Salah, L.; Benarous, K.; Linani, A.; Rabhi, F.; Chaib, K.; Chine, I.; Bensaidane, H.; Yousfi, M. Anti-inflammatory drugs as new inhibitors to xanthine oxidase: In vitro and in silico approach. Mol. Cell. Probes 2021, 58, 101733. [Google Scholar] [CrossRef]
- Bou-Salah, L.; Benarous, K.; Linani, A.; Bombarda, I.; Yousfi, M. In vitro and in silico inhibition studies of five essential oils on both enzymes human and bovine xanthine oxidase. Ind. Crops Prod. 2020, 143, 111949. [Google Scholar] [CrossRef]
- Benarous, K.; Bou-Salah, L.; Linani, A.; Yousfi, M.; Kostova, I.; Saso, L. Lanthanide (III) complexes of bis-coumarins as strong inhibitors of bovine xanthine oxidase-molecular docking and SAR studies. J. Biomol. Struct. Dyn. 2020, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Shintani, H. Determination of xanthine oxidase. Pharm. Anal. Acta 2013, 7, 4. [Google Scholar] [CrossRef] [Green Version]
- Jones, G.; Willett, P.; Glen, R.C.; Leach, A.R.; Taylor, R. Development and validation of a genetic algorithm for flexible docking. J. Mol. Biol. 1997, 267, 727–748. [Google Scholar] [CrossRef] [Green Version]
- HyperChem(TM) Professional 8.0; Hypercube, Inc.: Gainesville, FL, USA, 2002.
- Khedidja, B.; Abderrahman, L. Selection of orlistat as a potential inhibitor for lipase from Candida species. Bioinformation 2011, 7, 125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dassault Systèmes BIOVIA, BIOVIA Workbook, Release 2017; BIOVIA Pipeline Pilot—Dassault Systèmes: San Diego, CA, USA, 2017.
- Korb, O.; Stutzle, T.; Exner, T.E. Empirical scoring functions for advanced protein−ligand docking with PLANTS. J. Chem. Inf. Model. 2009, 49, 84–96. [Google Scholar] [CrossRef] [PubMed]
- Jones, G.; Willett, P.; Glen, R.C. Molecular recognition of receptor sites using a genetic algorithm with a description of desolvation. J. Mol. Biol. 1995, 245, 43–53. [Google Scholar] [CrossRef]
- Verdonk, M.L.; Cole, J.C.; Hartshorn, M.J.; Murray, C.W.; Taylor, R.D. Improved protein–ligand docking using GOLD. Proteins 2003, 52, 609–623. [Google Scholar] [CrossRef]






| IR | Raman | Vibrational Assignment | ||||
|---|---|---|---|---|---|---|
| HAOA | LaAOA | GaAOA | HAOA | LaAOA | GaAOA | |
| 3457 s | 3479 w | 3443 w | 3479 w | 3478 vw | ν(N1H1) | |
| 3361 m | 3355 m | 3456 vw | 3357 w | 3358 w | νas(NH2) | |
| 3333 s | 3448 w | 3336 m | 3323 m | 3334 w | ν(N3H3), ν(C-H) | |
| 3196 m | 3171 m | 3168 m | 3166 vw | νs(NH2) | ||
| 1691 vs | 1718 m | 1717 m | 1698 m | δ(NH); νs(C2=O2), ν(N–C6) | ||
| 1684 vs | 1691 vs | νs(C4=O4) | ||||
| 1667 s | 1673 vs | 1676 vs | 1678 sh | 1673 m | 1682 m | νs(C4=O4); ν(COO−), ν(C5=C6), δ(N3–H3) |
| 1604 s | 1637 vs | 1645 vs | 1612 vs | 1623 vs | 1628 vs | ν(C5=C6); β(NH2), ν(COO−) |
| 1566 m | 1556 m | 1553 m | 1560 m | 1542 w/m | 1546 w/m | δip(N1H1, N5H5); ν(C5C6), βs(NH2) |
| 1511 w | 1499 m | 1498 m | 1492 w/m | 1494 w | 1501 w | δ(NC); ν(ring); δip(N3H3) |
| 1457 m | 1447 w | 1433 w | ν(ring), βs(NH2), δ(Nl–H1), ν(COO−) | |||
| 1436 m | 1424 s | 1425 s | 1421 w | 1420 vs | 1407 vs | δ(N3H3), δ(ring), δ(N1H1) |
| 1405 m | 1390 s | 1391 s | 1384 s | 1388 s | δ(N3H3), δ(ring), δ(N1H1), νs(COO−) | |
| 1312 m | 1306 m | 1306 m | 1301 w | 1295 m/s | 1302 m/s | ν(C5–N), ν(C–N), δ(OH), βs(NH2) |
| 1255 m/s | 1290 sh | 1284 sh | ν(C–N), δ(N1H1), r(NH2), δ(ring) | |||
| 1234 m/s | 1237 w | 1239 w | 1242 m | 1230 m/s | 1240 m/s | ν(C–N), δ(N3H3), r(NH2), δ(NlH1) |
| 1140 sh | 1122 vw | 1126 vw | 1124 vw | 1131 vw | δ(OH) | |
| 1083 vw | 1047 vw | 1041 w | 1044 w | ν(C6-O, C6-C7), βas(NH2) | ||
| 989 sh | ν(NCN), δ(N3H3), r(NH2), δ(NlH1) | |||||
| 924 w | 941 sh | 944 sh | 919 w/m | 933 w/m | 948 w/m | ν(NCC), ν(ring), r(NH2), ν(COO) |
| 871 w/m | 884 vw | 875 vw | γ(N3–H3), γ(ring) | |||
| 795 vw | 798 m/807 | 797 m/807 | 809 vw | 788 sh | 781 sh | δop(O3C7O1) |
| 767 vw | 775 w | 777 w | 763 m | 777 m | γ(C4=O4), γ(C4–C=C6), γ(C6–C12) | |
| 754 w | 759 w | 759 w | 749 vw | γ(C6–C12), γ(C4=O4), γ(COOH), γ(N3–H) | ||
| 740 w | 747 w | 749 w | γ(C2=O2), γ(NC2N), γ(N3–H) | |||
| 696 vw | 694 vw | 692 vw | δ(ring), ΔsCOO), r(NH2) | |||
| 617 vw | 602 vw | ν(M-O) | ||||
| 584 vw | 595 vw | 582 w | 581 w/m | 589 w/m | δ(ring), Δs(COO) | |
| 521 sh | 509 sh | ν(M-O) | ||||
| 488 w | 501 w | 482 w/m | 480 m | 495 m | δ(ring), δ(NH2), Δs(COO) | |
| 474 w/m | 459 sh | 465 sh | γ(OH) | |||
| 446 m | 441 m | 442 m | 445 w | δ(OCNCO), δ(COO) + r(NH2) | ||
| 422 sh | 422 sh | 425 sh | 425 w/m | 438 w/m | τ(C2O2, ring), Δas(COO) | |
| 381 vw | 375 w | 374 w | δ(OCCN11), δ(COO), δ(C2=O), r(NH2); ν(M-O) | |||
| 249 w | 339 sh | 243 w | τ(NH2), Δs(COO) | |||
| 224 w | 205 w | ν(O-M-O) | ||||
| 196 w | 192 sh | 182 sh | τ(ring); δ(O-M-O) | |||
| Nn-H | HAOA | LaAOA | GaAOA |
|---|---|---|---|
| N1-H | 11.47 | 11.22 | 11.46 |
| N3-H | 9.44 | 9.03 | 8.48 |
| C5NH2-2H | 6.00 | 5.57 | 5.33 |
| Rank | Inhibitors | RR (%) | PLPChem Score | Nucleophilic Residues | Interaction Type | Length (Å) | Number of Interations | Fav/Unfav Bond |
|---|---|---|---|---|---|---|---|---|
| Control | Allopurinol | 100 | 23.89 | GLY798 | Hydrogen Bond | 2.15 | 04 | 8/0 |
| Hydrophobic Bond | 4.07 | 04 | ||||||
| 1 | LaAOA | 100 | 34.42 | GLN768 | Hydrogen Bond | 1.30 | 14 | 20/4 |
| Hydrophobic Bond | 4.24 | 05 | ||||||
| Coulombic | 3.47 | 01 | ||||||
| 2 | GaAOA | 90 | 33.35 | GLN1195 | Hydrogen Bond | 1.90 | 12 | 15/01 |
| Hydrophobic Bond | 4.46 | 03 | ||||||
| Coulombic | / | 00 | ||||||
| 3 | HAOA | 100 | 16.45 | ARG913 | Hydrogen Bond | 3.15 | 01 | 3/0 |
| Hydrophobic Bond | 4.38 | 02 | ||||||
| Coulombic | / | 00 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Todorov, L.; Saso, L.; Benarous, K.; Traykova, M.; Linani, A.; Kostova, I. Synthesis, Structure and Impact of 5-Aminoorotic Acid and Its Complexes with Lanthanum(III) and Gallium(III) on the Activity of Xanthine Oxidase. Molecules 2021, 26, 4503. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules26154503
Todorov L, Saso L, Benarous K, Traykova M, Linani A, Kostova I. Synthesis, Structure and Impact of 5-Aminoorotic Acid and Its Complexes with Lanthanum(III) and Gallium(III) on the Activity of Xanthine Oxidase. Molecules. 2021; 26(15):4503. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules26154503
Chicago/Turabian StyleTodorov, Lozan, Luciano Saso, Khedidja Benarous, Maria Traykova, Abderahmane Linani, and Irena Kostova. 2021. "Synthesis, Structure and Impact of 5-Aminoorotic Acid and Its Complexes with Lanthanum(III) and Gallium(III) on the Activity of Xanthine Oxidase" Molecules 26, no. 15: 4503. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules26154503