Natural Peptides Inducing Cancer Cell Death: Mechanisms and Properties of Specific Candidates for Cancer Therapeutics
Abstract
:1. Introduction
Methodology Used in Literature Research
2. Properties of Therapeutic Anticancer Peptides
2.1. Amino Acid Composition
2.2. Amphipathicity
2.3. Hydrophobicity
2.4. Net Charge
2.5. Secondary Structure in Membrane
2.6. Spatial Structure
2.7. Oligomerization Ability
3. Characteristics of Cancerous Cells Making Them Susceptible to Peptides
3.1. Negative Charge
3.2. Cholesterol Content
3.3. Microvilli
4. Cell Death of Cancerous Cells Mediated by Peptides
4.1. Disruption of Cell Membrane
4.1.1. Transient Pore Formation
4.1.2. Membrane Disruption
4.2. Necrosis and Apoptosis
4.2.1. Necrosis
4.2.2. Apoptosis
Extrinsic Pathway
Intrinsic Pathway
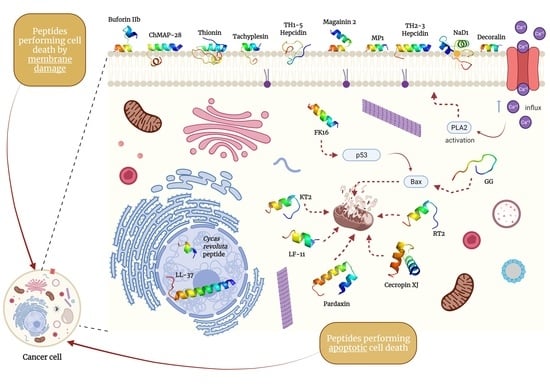
5. Specific Candidate Peptides as Anticancer Therapeutics
5.1. Peptides Performing Membrane-Damaging Cell Death
| Key | Peptide | Amino Acid Sequence | Reference |
|---|---|---|---|
| a | Buforin IIb | TRSSRAGLQFPVGRVHRLLRK | [96] |
| b | ChMAP-28 | GRFKRFRKKLKRLWHKVGPFVGPILHY | [97] |
| c | Decoralin | SLLSLIRKLIT | [98] |
| d & e | Hepcidin isoforms TH1-5 and TH2-3 | GIKCRFCCGCCTPGICGVCCRF & QSHLSLCRWCCNCCRSNKGC | [99,100] |
| f | Magainin 2 | GIGKFLHSAKKFGKAFVGEIMNS | [101] |
| g | NaD1 defensin | ARECKTESNTFPGICITKPPCRKACISEKFTDGHCSKILRRCLCTKPC | [102] |
| h | MP1 | ILGTILGLLKSL | [103] |
| i | Tachyplesin | KWCFRVCYRGICYRRCR | [104] |
| j | Thionin | KSCCRNTWARNCYNVCRLPGTISREICAKKCDCKIISGTTCPSDYPK | [105] |
5.1.1. Buforin IIb
5.1.2. ChMAP-28
5.1.3. Decoralin-NH2
5.1.4. Hepcidin
5.1.5. Magainin 2
5.1.6. NaD1 Defensin
5.1.7. MP1 Peptide
5.1.8. Tachyplesin
5.1.9. Thionins
5.2. Peptides Performing Apoptotic Cell Death
| Key | Peptide | Amino Acid Sequence | Reference |
|---|---|---|---|
| a | Cecropin XJ | WKIFKKIEKMGRNIRDGIVKAGPAIEVLGSAKAIGK | [146] |
| b | Cycas revoluta peptide | AWKLFDDGV | [147] |
| c | GG | GPPPQGGRPQG | [148] |
| d | LF11 | FQWQRNMRKVR | [149] |
| e & f | Leucrocins KT2 & RT2 | NGVQPKYKWWKWWKKWW & NGVQPKYRWWRWWRRWW | [150] |
| g & h | LL-37 native and its FK-16 fragment | FRKSKEKIGKEFKRIVQRIKDFLRNLVPRTES & FKRIVQRIKDFLRNLV | [151,152] |
| i | Pardaxin | GFFALIPKIISSPLFKTLLSAVGSALSSSGGQE | [153] |
5.2.1. Cecropin XJ
5.2.2. Cycas revoluta Peptide
5.2.3. GG Peptide
5.2.4. LF11 from Human Lactoferricin (hLFcin)
5.2.5. Leucrocins
5.2.6. LL-37 and Its FK-16 Fragment
5.2.7. Pardaxin
6. In Vivo Studies of Natural Specific Peptides
7. Disadvantages of Peptides Targeting Cancer Cells
8. Discussion
9. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mutgan, A.C.; Besikcioglu, H.E.; Wang, S.; Friess, H.; Ceyhan, G.O.; Demir, I.E. Insulin/IGF-driven cancer cell-stroma crosstalk as a novel therapeutic target in pancreatic cancer. Mol. Cancer 2018, 17, 66. [Google Scholar] [CrossRef] [Green Version]
- Soukup, T.; Lamb, B.W.; Arora, S.; Darzi, A.; Sevdalis, N.; Green, J.S.A. Successful strategies in implementing a multidisciplinary team working in the care of patients with cancer: An overview and synthesis of the available literature. J. Multidiscip. Healthc. 2018, 11, 49–61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, K.D.; Nogueira, L.; Mariotto, A.B.; Rowland, J.H.; Yabroff, K.R.; Alfano, C.M.; Jemal, A.; Kramer, J.L.; Siegel, R.L. Cancer treatment and survivorship statistics, 2019. CA Cancer J. Clin. 2019, 69, 363–385. [Google Scholar] [CrossRef] [Green Version]
- Hoda, M. Potential Alternatives to Conventional Cancer Therapeutic Approaches: The Way Forward. Curr. Pharm. Biotechnol. 2021, 22, 1141–1148. [Google Scholar] [CrossRef]
- Dallavalle, S.; Dobričić, V.; Lazzarato, L.; Gazzano, E.; Machuqueiro, M.; Pajeva, I.; Tsakovska, I.; Zidar, N.; Fruttero, R. Improvement of conventional anti-cancer drugs as new tools against multidrug resistant tumors. Drug Resist. Updat. 2020, 50, 100682. [Google Scholar] [CrossRef] [PubMed]
- Cui, W.; Aouidate, A.; Wang, S.; Yu, Q.; Li, Y.; Yuan, S. Discovering Anti-Cancer Drugs via Computational Methods. Front. Pharmacol. 2020, 11, 733. [Google Scholar] [CrossRef]
- Eghtedari, M.; Jafari Porzani, S.; Nowruzi, B. Anticancer potential of natural peptides from terrestrial and marine environments: A review. Phytochem. Lett. 2021, 42, 87–103. [Google Scholar] [CrossRef]
- Li, M.; Zhao, X.; Dai, J.; Yu, Z. Peptide therapeutics and assemblies for cancer immunotherapy. Sci. China Mater. 2019, 62, 1759–1781. [Google Scholar] [CrossRef] [Green Version]
- Tyagi, A.; Tuknait, A.; Anand, P.; Gupta, S.; Sharma, M.; Mathur, D.; Joshi, A.; Singh, S.; Gautam, A.; Raghava, G.P.S. CancerPPD: A database of anticancer peptides and proteins. Nucleic Acids Res. 2015, 43, D837–D843. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, S.; Chaudhary, K.; Dhanda, S.K.; Bhalla, S.; Usmani, S.S.; Gautam, A.; Tuknait, A.; Agrawal, P.; Mathur, D.; Raghava, G.P.S. SATPdb: A database of structurally annotated therapeutic peptides. Nucleic Acids Res. 2015, 44, D1119–D1126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Usmani, S.S.; Bedi, G.; Samuel, J.S.; Singh, S.; Kalra, S.; Kumar, P.; Ahuja, A.A.; Sharma, M.; Gautam, A.; Raghava, G.P.S. THPdb: Database of FDA-approved peptide and protein therapeutics. PLoS ONE 2017, 12, e0181748. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, W.; Ding, H.; Feng, P.; Lin, H.; Chou, K.-C. iACP: A sequence-based tool for identifying anticancer peptides. Oncotarget 2016, 7, 16895. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boopathi, V.; Subramaniyam, S.; Malik, A.; Lee, G.; Manavalan, B.; Yang, D.C. MACppred: A support vector machine-based meta-predictor for identification of anticancer peptides. Int. J. Mol. Sci. 2019, 20, 1964. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, G.; Li, X.; Wang, Z. APD2: The updated antimicrobial peptide database and its application in peptide design. Nucleic Acids Res. 2009, 37, 933–937. [Google Scholar] [CrossRef] [Green Version]
- Guzmán-Rodríguez, J.J.; Ochoa-Zarzosa, A.; López-Gómez, R.; López-Meza, J.E. Plant antimicrobial peptides as potential anticancer agents. Biomed Res. Int. 2015, 2015, 735087. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, L. Regulatory Considerations for Peptide Therapeutics. In Peptide Therapeutics: Strategy and Tactics for Chemistry, Manufacturing, and Controls; Royal Society of Chemistry: Cambridge, UK, 2019; pp. 1–30. ISBN 978-1-78801-644-5. [Google Scholar]
- D’Aloisio, V.; Dognini, P.; Hutcheon, G.A.; Coxon, C.R. PepTherDia: Database and structural composition analysis of approved peptide therapeutics and diagnostics. Drug Discov. Today 2021, 26, 1409–1419. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Zhang, C.; Chen, H.; Xue, J.; Guo, X.; Liang, M.; Chen, M. BioPepDB: An integrated data platform for food-derived bioactive peptides. Int. J. Food Sci. Nutr. 2018, 69, 963–968. [Google Scholar] [CrossRef] [PubMed]
- Farsinejad, S.; Gheisary, Z.; Ebrahimi Samani, S.; Alizadeh, A.M. Mitochondrial targeted peptides for cancer therapy. Tumor Biol. 2015, 36, 5715–5725. [Google Scholar] [CrossRef]
- Li, Y.; Yu, J. Research Progress in Structure-Activity Relationship of Bioactive Peptides. J. Med. Food 2015, 18, 147–156. [Google Scholar] [CrossRef]
- Fosgerau, K.; Hoffmann, T. Peptide therapeutics: Current status and future directions. Drug Discov. Today 2015, 20, 122–128. [Google Scholar] [CrossRef] [Green Version]
- Karami, Z.; Akbari-Adergani, B. Bioactive food derived peptides: A review on correlation between structure of bioactive peptides and their functional properties. J. Food Sci. Technol. 2019, 56, 535–547. [Google Scholar] [CrossRef] [PubMed]
- Habault, J.; Poyet, J.-L. Recent Advances in Cell Penetrating Peptide-Based Anticancer Therapies. Molecules 2019, 24, 927. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, H.; Choi, M.-C.; Seo, C.; Park, Y. Therapeutic Properties and Biological Benefits of Marine-Derived Anticancer Peptides. Int. J. Mol. Sci. 2018, 19, 919. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dang, T.; Süssmuth, R.D. Bioactive Peptide Natural Products as Lead Structures for Medicinal Use. Acc. Chem. Res. 2017, 50, 1566–1576. [Google Scholar] [CrossRef] [PubMed]
- Mishra, A.; Gauri, S.S.; Mukhopadhyay, S.K.; Chatterjee, S.; Das, S.S.; Mandal, S.M.; Dey, S. Identification and structural characterization of a new pro-apoptotic cyclic octapeptide cyclosaplin from somatic seedlings of Santalum album L. Peptides 2014, 54, 148–158. [Google Scholar] [CrossRef] [PubMed]
- Sarojini, V.; Cameron, A.J.; Varnava, K.G.; Denny, W.A.; Sanjayan, G. Cyclic Tetrapeptides from Nature and Design: A Review of Synthetic Methodologies, Structure, and Function. Chem. Rev. 2019. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Vorobyov, I.; Allen, T.W. The different interactions of lysine and arginine side chains with lipid membranes. J. Phys. Chem. B 2013, 117, 11906–11920. [Google Scholar] [CrossRef]
- Armstrong, C.T.; Mason, P.E.; Anderson, J.L.R.; Dempsey, C.E. Arginine side chain interactions and the role of arginine as a gating charge carrier in voltage sensitive ion channels. Sci. Rep. 2016, 6, 21759. [Google Scholar] [CrossRef] [Green Version]
- Kang, M.H.; Park, M.J.; Yoo, H.J.; Hyuk, K.Y.; Lee, S.G.; Kim, S.R.; Yeom, D.W.; Kang, M.J.; Choi, Y.W. RIPL peptide (IPLVVPLRRRRRRRRC)-conjugated liposomes for enhanced intracellular drug delivery to hepsin-expressing cancer cells. Eur. J. Pharm. Biopharm. 2014, 87, 489–499. [Google Scholar] [CrossRef]
- Sultana, A.; Luo, H.; Ramakrishna, S. Antimicrobial peptides and their applications in biomedical sector. Antibiotics 2021, 10, 1094. [Google Scholar] [CrossRef]
- Schaduangrat, N.; Nantasenamat, C.; Prachayasittikul, V.; Shoombuatong, W. ACPred: A Computational Tool for the Prediction and Analysis of Anticancer Peptides. Molecules 2019, 24, 1973. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.; Feng, Q.; Yan, Q.; Hao, X.; Chen, Y. Alpha-Helical Cationic Anticancer Peptides: A Promising Candidate for Novel Anticancer Drugs. Mini-Rev. Med. Chem. 2015, 15, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Gabernet, G.; Gautschi, D.; Müller, A.T.; Neuhaus, C.S.; Armbrecht, L.; Dittrich, P.S.; Hiss, J.A.; Schneider, G. In silico design and optimization of selective membranolytic anticancer peptides. Sci. Rep. 2019, 9, 11282. [Google Scholar] [CrossRef] [PubMed]
- Ditzinger, F.; Price, D.J.; Ilie, A.R.; Köhl, N.J.; Jankovic, S.; Tsakiridou, G.; Aleandri, S.; Kalantzi, L.; Holm, R.; Nair, A.; et al. Lipophilicity and hydrophobicity considerations in bio-enabling oral formulations approaches—A pearrl review. J. Pharm. Pharmacol. 2019, 71, 464–482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oelkrug, C.; Hartke, M.; Schubert, A. Mode of action of anticancer peptides (ACPs) from amphibian origin. Anticancer Res. 2015, 35, 635–643. [Google Scholar]
- Gabernet, G.; Müller, A.T.; Hiss, J.A.; Schneider, G. Membranolytic anticancer peptides. MedChemComm 2016, 7, 2232–2245. [Google Scholar] [CrossRef]
- Sun, X.; Acquah, C.; Aluko, R.E.; Udenigwe, C.C. Considering food matrix and gastrointestinal effects in enhancing bioactive peptide absorption and bioavailability. J. Funct. Foods 2020, 64, 103680. [Google Scholar] [CrossRef]
- Smolarczyk, T.; Roterman-Konieczna, I.; Stapor, K. Protein Secondary Structure Prediction: A Review of Progress and Directions. Curr. Bioinform. 2019, 15, 90–107. [Google Scholar] [CrossRef]
- Ma, Y.; Wu, Y.; Li, L. Relationship between primary structure or spatial conformation and functional activity of antioxidant peptides from Pinctada fucata. Food Chem. 2018, 264, 108–117. [Google Scholar] [CrossRef] [PubMed]
- Sah, B.N.P.; Vasiljevic, T.; McKechnie, S.; Donkor, O.N. Identification of Anticancer Peptides from Bovine Milk Proteins and Their Potential Roles in Management of Cancer: A Critical Review. Compr. Rev. Food Sci. Food Saf. 2015, 14, 123–138. [Google Scholar] [CrossRef]
- Janairo, J.I.B.; Janairo, G.C. Predicting Peptide Oligomeric State through Chemical Artificial Intelligence. Int. J. Pept. Res. Ther. 2021, 27, 763–767. [Google Scholar] [CrossRef]
- Vargas Casanova, Y.; Rodríguez Guerra, J.A.; Umaña Pérez, Y.A.; Leal Castro, A.L.; Almanzar Reina, G.; García Castañeda, J.E.; Rivera Monroy, Z.J. Antibacterial Synthetic Peptides Derived from Bovine Lactoferricin Exhibit Cytotoxic Effect against MDA-MB-468 and MDA-MB-231 Breast Cancer Cell Lines. Molecules 2017, 22, 1641. [Google Scholar] [CrossRef] [PubMed]
- Nyström, L.; Malmsten, M. Membrane interactions and cell selectivity of amphiphilic anticancer peptides. Curr. Opin. Colloid Interface Sci. 2018, 38, 1–7. [Google Scholar] [CrossRef]
- Tan, J.; Tay, J.; Hedrick, J.; Yang, Y.Y. Synthetic macromolecules as therapeutics that overcome resistance in cancer and microbial infection. Biomaterials 2020, 252, 120078. [Google Scholar] [CrossRef] [PubMed]
- Vitale, I.; Yamazaki, T.; Wennerberg, E.; Sveinbjørnsson, B.; Rekdal, Ø.; Demaria, S.; Galluzzi, L. Targeting Cancer Heterogeneity with Immune Responses Driven by Oncolytic Peptides. Trends Cancer 2021, 7, 557–572. [Google Scholar] [CrossRef] [PubMed]
- Kaushik, A.C.; Mehmood, A.; Peng, S.; Zhang, Y.J.; Dai, X.; Wei, D.Q. A-CaMP: A tool for anti-cancer and antimicrobial peptide generation. J. Biomol. Struct. Dyn. 2020, 39, 285–293. [Google Scholar] [CrossRef] [PubMed]
- Sharma, B.; Kanwar, S.S. Phosphatidylserine: A cancer cell targeting biomarker. Semin. Cancer Biol. 2018, 52, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Piotrowska, U.; Sobczak, M.; Oledzka, E. Current state of a dual behaviour of antimicrobial peptides—Therapeutic agents and promising delivery vectors. Chem. Biol. Drug Des. 2017, 90, 1079–1093. [Google Scholar] [CrossRef]
- Tornesello, A.L.; Borrelli, A.; Buonaguro, L.; Buonaguro, F.M.; Tornesello, M.L. Antimicrobial Peptides as Anticancer Agents: Functional Properties and Biological Activities. Molecules 2020, 25, 2850. [Google Scholar] [CrossRef]
- Leuschner, C.; Hansel, W. Membrane Disrupting Lytic Peptides for Cancer Treatments. Curr. Pharm. Des. 2005, 10, 2299–2310. [Google Scholar] [CrossRef]
- Baxter, A.A.; Lay, F.T.; Poon, I.K.H.; Kvansakul, M.; Hulett, M.D. Tumor cell membrane-targeting cationic antimicrobial peptides: Novel insights into mechanisms of action and therapeutic prospects. Cell. Mol. Life Sci. 2017, 74, 3809–3825. [Google Scholar] [CrossRef]
- Felício, M.R.; Silva, O.N.; Gonçalves, S.; Santos, N.C.; Franco, O.L. Peptides with Dual Antimicrobial and Anticancer Activities. Front. Chem. 2017, 5, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kunda, N.K. Antimicrobial peptides as novel therapeutics for non-small cell lung cancer. Drug Discov. Today 2020, 25, 238–247. [Google Scholar] [CrossRef] [PubMed]
- Ciumac, D.; Gong, H.; Hu, X.; Lu, J.R. Membrane targeting cationic antimicrobial peptides. J. Colloid Interface Sci. 2019, 537, 163–185. [Google Scholar] [CrossRef] [PubMed]
- Izzi, V.; Heljasvaara, R.; Pihlajaniemi, T. Understanding the extracellular matrix in acute myeloid leukemia. Haematologica 2017, 102, 1807–1809. [Google Scholar] [CrossRef]
- Chan, S.C.; Hui, L.; Chen, H.M. Enhancement of the cytolytic effect of anti-bacterial cecropin by the microvilli of cancer cells. Anticancer Res. 1998, 18, 4467–4474. [Google Scholar]
- Garizo, A.R.; Coelho, L.F.; Pinto, S.; Dias, T.P.; Fernandes, F.; Bernardes, N.; Fialho, A.M. The azurin-derived peptide ct-p19lc exhibits membrane-active properties and induces cancer cell death. Biomedicines 2021, 9, 1194. [Google Scholar] [CrossRef]
- Liao, W.; Lai, T.; Chen, L.; Fu, J.; Sreenivasan, S.T.; Yu, Z.; Ren, J. Synthesis and Characterization of a Walnut Peptides-Zinc Complex and Its Antiproliferative Activity against Human Breast Carcinoma Cells through the Induction of Apoptosis. J. Agric. Food Chem. 2016, 64, 1509–1519. [Google Scholar] [CrossRef]
- Riedl, S.; Leber, R.; Rinner, B.; Schaider, H.; Lohner, K.; Zweytick, D. Human lactoferricin derived di-peptides deploying loop structures induce apoptosis specifically in cancer cells through targeting membranous phosphatidylserine. Biochim. Biophys. Acta—Biomembr. 2015, 1848, 2918–2931. [Google Scholar] [CrossRef] [Green Version]
- Gaspar, D.; Salomé Veiga, A.; Castanho, M.A.R.B. From antimicrobial to anticancer peptides. A review. Front. Microbiol. 2013, 4, 294. [Google Scholar] [CrossRef] [Green Version]
- Mesa-Galloso, H.; Pedrera, L.; Ros, U. Pore-forming proteins: From defense factors to endogenous executors of cell death. Chem. Phys. Lipids 2021, 234, 105026. [Google Scholar] [CrossRef] [PubMed]
- Lipkin, R.; Lazaridis, T. Computational studies of peptide-induced membrane pore formation. Philos. Trans. R. Soc. B Biol. Sci. 2017, 372, 20160219. [Google Scholar] [CrossRef] [Green Version]
- Sepehri, A.; PeBenito, L.; Pino-Angeles, A.; Lazaridis, T. What Makes a Good Pore Former: A Study of Synthetic Melittin Derivatives. Biophys. J. 2020, 118, 1901–1913. [Google Scholar] [CrossRef]
- Wimley, W.C.; Hristova, K. The Mechanism of Membrane Permeabilization by Peptides: Still an Enigma. Aust. J. Chem. 2020, 73, 96–103. [Google Scholar] [CrossRef]
- Pitsalidis, C.; Pappa, A.M.; Porel, M.; Artim, C.M.; Faria, G.C.; Duong, D.D.; Alabi, C.A.; Daniel, S.; Salleo, A.; Owens, R.M. Biomimetic Electronic Devices for Measuring Bacterial Membrane Disruption. Adv. Mater. 2018, 30, 1803130. [Google Scholar] [CrossRef] [PubMed]
- Togo, T. Autocrine purinergic signaling stimulated by cell membrane disruption is involved in both cell membrane repair and adaptive response in MDCK cells. Biochem. Biophys. Res. Commun. 2019, 511, 161–164. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Li, Y.; Li, Z.; Lan, X.; Leung, P.H.-M.; Li, J.; Yang, M.; Ko, F.; Qin, L. Mechanism of Anticancer Effects of Antimicrobial Peptides. J. Fiber Bioeng. Inform. 2015, 8, 25–36. [Google Scholar] [CrossRef]
- Dennison, S.R.; Harris, F.; Phoenix, D.A. Investigations into the potential anticancer activity of Maximin H5. Biochimie 2017, 137, 29–34. [Google Scholar] [CrossRef] [Green Version]
- Sekiya, Y.; Sakashita, S.; Shimizu, K.; Usui, K.; Kawano, R. Channel current analysis estimates the pore-formation and the penetration of transmembrane peptides. Analyst 2018, 143, 3540–3543. [Google Scholar] [CrossRef] [PubMed]
- Tang, D.; Kang, R.; Berghe, T.V.; Vandenabeele, P.; Kroemer, G. The molecular machinery of regulated cell death. Cell Res. 2019, 29, 347–364. [Google Scholar] [CrossRef] [Green Version]
- Braicu, C.; Zanoaga, O.; Zimta, A.A.; Tigu, A.B.; Kilpatrick, K.L.; Bishayee, A.; Nabavi, S.M.; Berindan-Neagoe, I. Natural compounds modulate the crosstalk between apoptosis- and autophagy-regulated signaling pathways: Controlling the uncontrolled expansion of tumor cells. Semin. Cancer Biol. 2020. [Google Scholar] [CrossRef] [PubMed]
- D’Arcy, M.S. Cell death: A review of the major forms of apoptosis, necrosis and autophagy. Cell Biol. Int. 2019, 43, 582–592. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Kang, J.; Fu, C. The independence of and associations among apoptosis, autophagy, and necrosis. Signal Transduct. Target. Ther. 2018, 3, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alharbi, K.S.; Fuloria, N.K.; Fuloria, S.; Rahman, S.B.; Al-Malki, W.H.; Javed Shaikh, M.A.; Thangavelu, L.; Singh, S.K.; Rama Raju Allam, V.S.; Jha, N.K.; et al. Nuclear factor-kappa B and its role in inflammatory lung disease. Chem. Biol. Interact. 2021, 345, 109568. [Google Scholar] [CrossRef]
- Tonnus, W.; Meyer, C.; Paliege, A.; Belavgeni, A.; von Mässenhausen, A.; Bornstein, S.R.; Hugo, C.; Becker, J.U.; Linkermann, A. The pathological features of regulated necrosis. J. Pathol. 2019, 247, 697–707. [Google Scholar] [CrossRef] [PubMed]
- Grootjans, S.; Vanden Berghe, T.; Vandenabeele, P. Initiation and execution mechanisms of necroptosis: An overview. Cell Death Differ. 2017, 24, 1184–1195. [Google Scholar] [CrossRef] [PubMed]
- Pfeffer, C.M.; Singh, A.T.K. Apoptosis: A target for anticancer therapy. Int. J. Mol. Sci. 2018, 19, 448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kesavardhana, S.; Malireddi, R.K.S.; Kanneganti, T.D. Caspases in Cell Death, Inflammation, and Pyroptosis. Annu. Rev. Immunol. 2020, 38, 567–595. [Google Scholar] [CrossRef] [Green Version]
- Mohammadinejad, R.; Moosavi, M.A.; Tavakol, S.; Vardar, D.Ö.; Hosseini, A.; Rahmati, M.; Dini, L.; Hussain, S.; Mandegary, A.; Klionsky, D.J. Necrotic, apoptotic and autophagic cell fates triggered by nanoparticles. Autophagy 2019, 15, 4–33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.; Yuan, W.; Lin, Z. Functional roles in cell signaling of adaptor protein TRADD from a structural perspective. Comput. Struct. Biotechnol. J. 2020, 18, 2867–2876. [Google Scholar] [CrossRef] [PubMed]
- Füllsack, S.; Rosenthal, A.; Wajant, H.; Siegmund, D. Redundant and receptor-specific activities of TRADD, RIPK1 and FADD in death receptor signaling. Cell Death Dis. 2019, 10, 122. [Google Scholar] [CrossRef] [PubMed]
- Mouasni, S.; Tourneur, L. FADD at the Crossroads between Cancer and Inflammation. Trends Immunol. 2018, 39, 1036–1053. [Google Scholar] [CrossRef] [PubMed]
- Ivanisenko, N.V.; Lavrik, I.N. Mechanisms of Procaspase-8 Activation in the Extrinsic Programmed Cell Death Pathway. Mol. Biol. 2019, 53, 732–738. [Google Scholar] [CrossRef]
- Holbrook, J.; Lara-Reyna, S.; Jarosz-Griffiths, H.; McDermott, M. Tumour necrosis factor signalling in health and disease [version 1; referees: 2 approved]. F1000Research 2019, 8. [Google Scholar] [CrossRef] [PubMed]
- Wallach, D. The tumor necrosis factor family: Family conventions and private idiosyncrasies. Cold Spring Harb. Perspect. Biol. 2018, 10, a028431. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.; Walker, A.J.; Berk, M.; Maes, M.; Puri, B.K. Cell Death Pathways: A Novel Therapeutic Approach for Neuroscientists. Mol. Neurobiol. 2018, 55, 5767–5786. [Google Scholar] [CrossRef] [Green Version]
- Liu, G.; Yuan, Y.; Long, M.; Luo, T.; Bian, J.; Liu, X.; Gu, J.; Zou, H.; Song, R.; Wang, Y.; et al. Beclin-1-mediated Autophagy Protects Against Cadmium-activated Apoptosis via the Fas/FasL Pathway in Primary Rat Proximal Tubular Cell Culture. Sci. Rep. 2017, 7, 977. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Warren, C.F.A.; Wong-Brown, M.W.; Bowden, N.A. BCL-2 family isoforms in apoptosis and cancer. Cell Death Dis. 2019, 10, 177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peña-Blanco, A.; García-Sáez, A.J. Bax, Bak and beyond—Mitochondrial performance in apoptosis. FEBS J. 2018, 285, 416–431. [Google Scholar] [CrossRef] [Green Version]
- Jan, R.; Chaudhry, G.-S. Understanding Apoptosis and Apoptotic Pathways Targeted Cancer Therapeutics. Adv. Pharm. Bull. 2019, 9, 205–218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shakeri, R.; Kheirollahi, A.; Davoodi, J. Apaf-1: Regulation and function in cell death. Biochimie 2017, 135, 111–125. [Google Scholar] [CrossRef]
- Wang, S.H.; Yu, J. Structure-based design for binding peptides in anti-cancer therapy. Biomaterials 2018, 156, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Gerlach, S.L.; Rathinakumar, R.; Chakravarty, G.; Göransson, U.; Wimley, W.C.; Darwin, S.P.; Mondal, D. Anticancer and chemosensitizing abilities of cycloviolacin O2 from Viola odorata and psyle cyclotides from Psychotria leptothyrsa. Biopolymers 2010, 94, 617–625. [Google Scholar] [CrossRef]
- Li, Y.; Xiang, Q.; Zhang, Q.; Huang, Y.; Su, Z. Overview on the recent study of antimicrobial peptides: Origins, functions, relative mechanisms and application. Peptides 2012, 37, 207–215. [Google Scholar] [CrossRef] [PubMed]
- Lan, Y.; Ye, Y.; Kozlowska, J.; Lam, J.K.W.; Drake, A.F.; Mason, A.J. Structural contributions to the intracellular targeting strategies of antimicrobial peptides. Biochim. Biophys. Acta—Biomembr. 2010, 1798, 1934–1943. [Google Scholar] [CrossRef] [Green Version]
- Emelianova, A.A.; Kuzmin, D.V.; Panteleev, P.V.; Sorokin, M.; Buzdin, A.A.; Ovchinnikova, T.V. Anticancer Activity of the Goat Antimicrobial Peptide ChMAP-28. Front. Pharmacol. 2018, 9, 1501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Konno, K.; Rangel, M.; Oliveira, J.S.; dos Santos Cabrera, M.P.; Fontana, R.; Hirata, I.Y.; Hide, I.; Nakata, Y.; Mori, K.; Kawano, M.; et al. Decoralin, a novel linear cationic α-helical peptide from the venom of the solitary eumenine wasp Oreumenes decoratus. Peptides 2007, 28, 2320–2327. [Google Scholar] [CrossRef] [PubMed]
- Lo, Y.L.; Lee, H.P.; Tu, W.C. The Use of a Liposomal Formulation Incorporating an Antimicrobial Peptide from Tilapia as a New Adjuvant to Epirubicin in Human Squamous Cell Carcinoma and Pluripotent Testicular Embryonic Carcinoma Cells. Int. J. Mol. Sci. 2015, 16, 22711–22734. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.Y.; Lin, W.J.; Lin, T.L. A fish antimicrobial peptide, tilapia hepcidin TH2-3, shows potent antitumor activity against human fibrosarcoma cells. Peptides 2009, 30, 1636–1642. [Google Scholar] [CrossRef] [PubMed]
- Aisenbrey, C.; Amaro, M.; Pospíšil, P.; Hof, M.; Bechinger, B. Highly synergistic antimicrobial activity of magainin 2 and PGLa peptides is rooted in the formation of supramolecular complexes with lipids. Sci. Rep. 2020, 10, 11652. [Google Scholar] [CrossRef]
- Järvå, M.; Lay, F.T.; Phan, T.K.; Humble, C.; Poon, I.K.H.; Bleackley, M.R.; Anderson, M.A.; Hulett, M.D.; Kvansakul, M. X-ray structure of a carpet-like antimicrobial defensin-phospholipid membrane disruption complex. Nat. Commun. 2018, 9, 1962. [Google Scholar] [CrossRef] [PubMed]
- Souza, B.M.; Mendes, M.A.; Santos, L.D.; Marques, M.R.; César, L.M.M.; Almeida, R.N.A.; Pagnocca, F.C.; Konno, K.; Palma, M.S. Structural and functional characterization of two novel peptide toxins isolated from the venom of the social wasp Polybia paulista. Peptides 2005, 26, 2157–2164. [Google Scholar] [CrossRef] [PubMed]
- Laederach, A.; Andreotti, A.H.; Bruce Fulton, D. Solution and Micelle-Bound Structures of Tachyplesin I and Its Active Aromatic Linear Derivatives. Biochemistry 2002, 41, 12359–12368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vila-Perelló, M.; Sánchez-Vallet, A.; García-Olmedo, F.; Molina, A.; Andreu, D. Synthetic and structural studies on Pyrularia pubera thionin: A single-residue mutation enhances activity against Gram-negative bacteria. FEBS Lett. 2003, 536, 215–219. [Google Scholar] [CrossRef] [Green Version]
- Cardoso, M.H.; Meneguetti, B.T.; Costa, B.O.; Buccini, D.F.; Oshiro, K.G.N.; Preza, S.L.E.; Carvalho, C.M.E.; Migliolo, L.; Franco, O.L. Non-lytic antibacterial peptides that translocate through bacterial membranes to act on intracellular targets. Int. J. Mol. Sci. 2019, 20, 4877. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, D.; Xu, Y. Buforin IIb induced cell cycle arrest in liver cancer. Anim. Cells Syst. 2019, 23, 176–183. [Google Scholar] [CrossRef]
- Liscano, Y.; Oñate-Garzón, J.; Delgado, J.P. Peptides with dual antimicrobial–anticancer activity: Strategies to overcome peptide limitations and rational design of anticancer peptides. Molecules 2020, 25, 4245. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Yang, N.; Teng, D.; Wang, X.; Mao, R.; Wang, J. A review of the design and modification of lactoferricins and their derivatives. BioMetals 2018, 31, 331–341. [Google Scholar] [CrossRef]
- Oliva Arguelles, B.; Riera-Romo, M.; Guerra Vallespi, M. Antitumour peptide based on a protein derived from the horseshoe crab: CIGB-552 a promising candidate for cancer therapy. Br. J. Pharmacol. 2020, 177, 3625–3634. [Google Scholar] [CrossRef] [PubMed]
- Abd El-Wahed, A.; Yosri, N.; Sakr, H.H.; Du, M.; Algethami, A.F.M.; Zhao, C.; Abdelazeem, A.H.; Tahir, H.E.; Masry, S.H.D.; Abdel-Daim, M.M.; et al. Wasp Venom Biochemical Components and Their Potential in Biological Applications and Nanotechnological Interventions. Toxins 2021, 13, 206. [Google Scholar] [CrossRef] [PubMed]
- Pillong, M.; Hiss, J.A.; Schneider, P.; Lin, Y.-C.; Posselt, G.; Pfeiffer, B.; Blatter, M.; Müller, A.T.; Bachler, S.; Neuhaus, C.S.; et al. Rational Design of Membrane-Pore-Forming Peptides. Small 2017, 13, 1701316. [Google Scholar] [CrossRef]
- Torres, M.D.T.; Andrade, G.P.; Sato, R.H.; Pedron, C.N.; Manieri, T.M.; Cerchiaro, G.; Ribeiro, A.O.; De la Fuente-Nunez, C.; Oliveira, V.X. Natural and redesigned wasp venom peptides with selective antitumoral activity. Beilstein J. Org. Chem. 2018, 14, 1693–1703. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Torres, M.D.T.; Pedron, C.N.; da Silva Lima, J.A.; da Silva, P.I.; da Silva, F.D.; Oliveira, V.X. Antimicrobial activity of leucine-substituted decoralin analogs with lower hemolytic activity. J. Pept. Sci. 2017, 23, 818–823. [Google Scholar] [CrossRef] [PubMed]
- Ting, C.H.; Pan, C.Y.; Chen, Y.C.; Lin, Y.C.; Chen, T.Y.; Rajanbabu, V.; Chen, J.Y. Impact of Tilapia hepcidin 2-3 dietary supplementation on the gut microbiota profile and immunomodulation in the grouper (Epinephelus lanceolatus). Sci. Rep. 2019, 9, 19047. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, W.T.; Pan, C.Y.; Rajanbabu, V.; Cheng, C.W.; Chen, J.Y. Tilapia (Oreochromis mossambicus) antimicrobial peptide, hepcidin 1–5, shows antitumor activity in cancer cells. Peptides 2011, 32, 342–352. [Google Scholar] [CrossRef]
- Kim, M.K.; Kang, N.; Ko, S.J.; Park, J.; Park, E.; Shin, D.W.; Kim, S.H.; Lee, S.A.; Lee, J.I.; Lee, S.H.; et al. Antibacterial and antibiofilm activity and mode of action of magainin 2 against drug-resistant acinetobacter baumannii. Int. J. Mol. Sci. 2018, 19, 3041. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zerweck, J.; Strandberg, E.; Kukharenko, O.; Reichert, J.; Bürck, J.; Wadhwani, P.; Ulrich, A.S. Molecular mechanism of synergy between the antimicrobial peptides PGLa and magainin 2. Sci. Rep. 2017, 7, 13153. [Google Scholar] [CrossRef]
- Rashid, M.M.O.; Moghal, M.M.R.; Billah, M.M.; Hasan, M.; Yamazaki, M. Effect of membrane potential on pore formation by the antimicrobial peptide magainin 2 in lipid bilayers. Biochim. Biophys. Acta—Biomembr. 2020, 1862, 183381. [Google Scholar] [CrossRef]
- Liu, S.; Yang, H.; Wan, L.; Cai, H.W.; Li, S.F.; Li, Y.P.; Cheng, J.Q.; Lu, X.F. Enhancement of cytotoxicity of antimicrobial peptide magainin II in tumor cells by bombesin-targeted delivery. Acta Pharmacol. Sin. 2011, 32, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Deslouches, B.; Peter Di, Y. Antimicrobial peptides with selective antitumor mechanisms: Prospect for anticancer applications. Oncotarget 2017, 8, 46635–46651. [Google Scholar] [CrossRef] [Green Version]
- De Oliveira Carvalho, A.; Moreira Gomes, V. Plant Defensins and Defensin-Like Peptides—Biological Activities and Biotechnological Applications. Curr. Pharm. Des. 2012, 17, 4270–4293. [Google Scholar] [CrossRef]
- Silva, P.M.; Gonçalves, S.; Santos, N.C. Defensins: Antifungal lessons from eukaryotes. Front. Microbiol. 2014, 5, 97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vriens, K.; Cammue, B.; Thevissen, K. Antifungal Plant Defensins: Mechanisms of Action and Production. Molecules 2014, 19, 12280–12303. [Google Scholar] [CrossRef] [Green Version]
- Phan, T.K.; Lay, F.T.; Poon, I.K.H.; Hinds, M.G.; Kvansakul, M.; Hulett, M.D. Human β-defensin 3 contains an oncolytic motif that binds PI(4,5)P2 to mediate tumour cell permeabilisation. Oncotarget 2016, 7, 2054–2069. [Google Scholar] [CrossRef] [Green Version]
- Shafee, T.M.A.; Lay, F.T.; Phan, T.K.; Anderson, M.A.; Hulett, M.D. Convergent evolution of defensin sequence, structure and function. Cell. Mol. Life Sci. 2017, 74, 663–682. [Google Scholar] [CrossRef]
- Baxter, A.A.; Poon, I.K.H.; Hulett, M.D. The plant defensin NaD1 induces tumor cell death via a non-apoptotic, membranolytic process. Cell Death Discov. 2017, 3, 16102. [Google Scholar] [CrossRef] [Green Version]
- Dos Santos Cabrera, M.P.; Arcisio-Miranda, M.; Gorjão, R.; Leite, N.B.; De Souza, B.M.; Curi, R.; Procopio, J.; Ruggiero Neto, J.; Palma, M.S. Influence of the bilayer composition on the binding and membrane disrupting effect of polybia-MP1, an antimicrobial mastoparan peptide with leukemic T-lymphocyte cell selectivity. Biochemistry 2012, 51, 4898–4908. [Google Scholar] [CrossRef] [PubMed]
- Leite, N.B.; Dos Santos Alvares, D.; De Souza, B.M.; Palma, M.S.; Ruggiero Neto, J. Effect of the aspartic acid D2 on the affinity of Polybia-MP1 to anionic lipid vesicles. Eur. Biophys. J. 2014, 43, 121–130. [Google Scholar] [CrossRef] [PubMed]
- Leite, N.B.; Aufderhorst-Roberts, A.; Palma, M.S.; Connell, S.D.; Neto, J.R.; Beales, P.A. PE and PS Lipids Synergistically Enhance Membrane Poration by a Peptide with Anticancer Properties. Biophys. J. 2015, 109, 936–947. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xuan, H.L.; Duc, T.D.; Thuy, A.M.; Chau, P.M.; Tung, T.T. Chemical approaches in the development of natural nontoxic peptide Polybia-MP1 as a potential dual antimicrobial and antitumor agent. Amino Acids 2021, 53, 843–852. [Google Scholar] [CrossRef]
- Alvares, D.S.; Ruggiero Neto, J.; Ambroggio, E.E. Phosphatidylserine lipids and membrane order precisely regulate the activity of Polybia-MP1 peptide. Biochim. Biophys. Acta—Biomembr. 2017, 1859, 1067–1074. [Google Scholar] [CrossRef] [PubMed]
- Alvares, D.S.; Fanani, M.L.; Ruggiero Neto, J.; Wilke, N. The interfacial properties of the peptide Polybia-MP1 and its interaction with DPPC are modulated by lateral electrostatic attractions. Biochim. Biophys. Acta—Biomembr. 2016, 1858, 393–402. [Google Scholar] [CrossRef] [PubMed]
- Alvares, D.S.; Wilke, N.; Ruggiero Neto, J.; Fanani, M.L. The insertion of Polybia-MP1 peptide into phospholipid monolayers is regulated by its anionic nature and phase state. Chem. Phys. Lipids 2017, 207, 38–48. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, A.M.B.; Silva-Gonçalves, L.C.; Oliveira, F.A.; Arcisio-Miranda, M. Pro-necrotic Activity of Cationic Mastoparan Peptides in Human Glioblastoma Multiforme Cells Via Membranolytic Action. Mol. Neurobiol. 2018, 55, 5490–5504. [Google Scholar] [CrossRef]
- Takahashi, H.; Yumoto, K.; Yasuhara, K.; Nadres, E.T.; Kikuchi, Y.; Taichman, R.S.; Kuroda, K. Anticancer polymers designed for killing dormant prostate cancer cells. Sci. Rep. 2019, 9, 1096. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vernen, F.; Harvey, P.J.; Dias, S.A.; Veiga, A.S.; Huang, Y.H.; Craik, D.J.; Lawrence, N.; Henriques, S.T. Characterization of tachyplesin peptides and their cyclized analogues to improve antimicrobial and anticancer properties. Int. J. Mol. Sci. 2019, 20, 4184. [Google Scholar] [CrossRef] [Green Version]
- Vernen, F.; Craik, D.J.; Lawrence, N.; Troeira Henriques, S. Cyclic Analogues of Horseshoe Crab Peptide Tachyplesin i with Anticancer and Cell Penetrating Properties. ACS Chem. Biol. 2019, 14, 2895–2908. [Google Scholar] [CrossRef]
- Liu, C.; Qi, J.; Shan, B.; Ma, Y. Tachyplesin causes membrane instability that kills multidrug-resistant bacteria by inhibiting the 3-ketoacyl carrier protein reductase FabG. Front. Microbiol. 2018, 9, 825. [Google Scholar] [CrossRef]
- Stec, B. Plant thionins—The structural perspective. Cell. Mol. Life Sci. 2006, 63, 1370–1385. [Google Scholar] [CrossRef]
- Prabhu, S.; Dennison, S.R.; Lea, B.; Snape, T.J.; Nicholl, I.D.; Radecka, I.; Harris, F. Anionic Antimicrobial and Anticancer Peptides from Plants. CRC. Crit. Rev. Plant Sci. 2013, 32, 303–320. [Google Scholar] [CrossRef]
- Gaspar, D.; Castanho, M.A.R.B. Anticancer peptides: Prospective innovation in cancer therapy. In Host Defense Peptides and Their Potential as Therapeutic Agents; Springer International Publishing: Cham, Switzerland, 2016; pp. 95–109. ISBN 9783319329499. [Google Scholar]
- Patel, S.; Akhtar, N. Antimicrobial peptides (AMPs): The quintessential ‘offense and defense’ molecules are more than antimicrobials. Biomed. Pharmacother. 2017, 95, 1276–1283. [Google Scholar] [CrossRef] [PubMed]
- Kaczanowski, S. Symbiotic Origin of Apoptosis. Results Probl. Cell Differ. 2020, 69, 253–280. [Google Scholar] [CrossRef] [PubMed]
- Min, K.A.; Maharjan, P.; Ham, S.; Shin, M.C. Pro-apoptotic peptides-based cancer therapies: Challenges and strategies to enhance therapeutic efficacy. Arch. Pharm. Res. 2018, 41, 594–616. [Google Scholar] [CrossRef]
- Xia, L.J.; Wu, Y.L.; Ma, J.; Zhang, F.C. Therapeutic effects of antimicrobial peptide on malignant ascites in a mouse model. Mol. Med. Rep. 2018, 17, 6245–6252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mandal, S.M.; Migliolo, L.; Das, S.; Mandal, M.; Franco, O.L.; Hazra, T.K. Identification and characterization of a bactericidal and proapoptotic peptide from cycas revoluta seeds with DNA binding properties. J. Cell. Biochem. 2012, 113, 184–193. [Google Scholar] [CrossRef]
- Trindade, F.; Amado, F.; Pinto da Costa, J.; Ferreira, R.; Maia, C.; Henriques, I.; Colaço, B.; Vitorino, R. Salivary peptidomic as a tool to disclose new potential antimicrobial peptides. J. Proteomics 2015, 115, 49–57. [Google Scholar] [CrossRef]
- Azuma, M.; Del Carpio, C.A.; Kojima, T.; Yokoyama, I.; Tajiri, H.; Yoshikawa, K.; Saga, S. Antibacterial activity of multiple antigen peptides homologous to a loop region in human lactoferrin. J. Pept. Res. 2008, 54, 237–241. [Google Scholar] [CrossRef]
- Anunthawan, T.; Yaraksa, N.; Phosri, S.; Theansungnoen, T.; Daduang, S.; Dhiravisit, A.; Thammasirirak, S. Improving the antibacterial activity and selectivity of an ultra short peptide by hydrophobic and hydrophilic amino acid stretches. Bioorg. Med. Chem. Lett. 2013, 23, 4657–4662. [Google Scholar] [CrossRef]
- Okumura, K.; Itoh, A.; Isogai, E.; Hirose, K.; Hosokawa, Y.; Abiko, Y.; Shibata, T.; Hirata, M.; Isogai, H. C-terminal domain of human CAP18 antimicrobial peptide induces apoptosis in oral squamous cell carcinoma SAS-H1 cells. Cancer Lett. 2004, 212, 185–194. [Google Scholar] [CrossRef]
- Ren, S.X.; Shen, J.; Cheng, A.S.L.; Lu, L.; Chan, R.L.Y.; Li, Z.J.; Wang, X.J.; Wong, C.C.M.; Zhang, L.; Ng, S.S.M.; et al. FK-16 Derived from the Anticancer Peptide LL-37 Induces Caspase-Independent Apoptosis and Autophagic Cell Death in Colon Cancer Cells. PLoS ONE 2013, 8, e063641. [Google Scholar] [CrossRef] [Green Version]
- Shai, Y.; Bach, D.; Yanovsky, A. Channel formation properties of synthetic pardaxin and analogues. J. Biol. Chem. 1990, 265, 20202–20209. [Google Scholar] [CrossRef]
- Xia, L.; Wu, Y.; Ma, J.; Yang, J.; Zhang, F. The antibacterial peptide from Bombyx mori cecropinXJ induced growth arrest and apoptosis in human hepatocellular carcinoma cells. Oncol. Lett. 2016, 12, 57–62. [Google Scholar] [CrossRef] [Green Version]
- Ramos-Martín, F.; D’Amelio, N. Molecular basis of the anticancer and antibacterial properties of cecropinxj peptide: An in silico study. Int. J. Mol. Sci. 2021, 22, 691. [Google Scholar] [CrossRef]
- Chang, V.H.S.; Yang, D.H.A.; Lin, H.H.; Pearce, G.; Ryan, C.A.; Chen, Y.C. IbACP, a sixteen-amino-acid peptide isolated from Ipomoea batatas leaves, induces carcinoma cell apoptosis. Peptides 2013, 47, 148–156. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, S.; Kato, K.; Koba, A.; Minami, Y.; Watanabe, K.; Yagi, F. Purification, characterization, and sequencing of antimicrobial peptides, Cy-AMP1, Cy-AMP2, and Cy-AMP3, from the Cycad (Cycas revoluta) seeds. Peptides 2008, 29, 2110–2117. [Google Scholar] [CrossRef]
- Barbosa Pelegrini, P.; Del Sarto, R.P.; Silva, O.N.; Franco, O.L.; Grossi-De-Sa, M.F. Antibacterial peptides from plants: What they are and how they probably work. Biochem. Res. Int. 2011, 2011, 250349. [Google Scholar] [CrossRef] [Green Version]
- Dijk, I.A.; Laura Ferrando, M.; Wijk, A.; Hoebe, R.A.; Nazmi, K.; Jonge, W.J.; Krawczyk, P.M.; Bolscher, J.G.M.; Veerman, E.C.I.; Stap, J. Human salivary peptide histatin-1 stimulates epithelial and endothelial cell adhesion and barrier function. FASEB J. 2017, 31, 3922–3933. [Google Scholar] [CrossRef] [Green Version]
- Grant, M.; Kilsgård, O.; Åkerman, S.; Klinge, B.; Demmer, R.T.; Malmström, J.; Jönsson, D. The Human Salivary Antimicrobial Peptide Profile according to the Oral Microbiota in Health, Periodontitis and Smoking. J. Innate Immun. 2019, 11, 432–444. [Google Scholar] [CrossRef]
- Da Costa, J.P.; Carvalhais, V.; Amado, F.; Silva, A.; Nogueira-Ferreira, R.; Ferreira, R.; Helguero, L.; Vitorino, R. Anti-tumoral activity of human salivary peptides. Peptides 2015, 71, 170–178. [Google Scholar] [CrossRef]
- Vila, T.; Rizk, A.M.; Sultan, A.S.; Jabra-Rizk, M.A. The power of saliva: Antimicrobial and beyond. PLoS Pathog. 2019, 15, e1008058. [Google Scholar] [CrossRef]
- Morici, P.; Florio, W.; Rizzato, C.; Ghelardi, E.; Tavanti, A.; Rossolini, G.M.; Lupetti, A. Synergistic activity of synthetic N-terminal peptide of human lactoferrin in combination with various antibiotics against carbapenem-resistant Klebsiella pneumoniae strains. Eur. J. Clin. Microbiol. Infect. Dis. 2017, 36, 1739–1748. [Google Scholar] [CrossRef]
- Zweytick, D.; Pabst, G.; Abuja, P.M.; Jilek, A.; Blondelle, S.E.; Andrä, J.; Jerala, R.; Monreal, D.; Martinez de Tejada, G.; Lohner, K. Influence of N-acylation of a peptide derived from human lactoferricin on membrane selectivity. Biochim. Biophys. Acta—Biomembr. 2006, 1758, 1426–1435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iglesias-Figueroa, B.F.; Siqueiros-Cendón, T.S.; Gutierrez, D.A.; Aguilera, R.J.; Espinoza-Sánchez, E.A.; Arévalo-Gallegos, S.; Varela-Ramirez, A.; Rascón-Cruz, Q. Recombinant human lactoferrin induces apoptosis, disruption of F-actin structure and cell cycle arrest with selective cytotoxicity on human triple negative breast cancer cells. Apoptosis 2019, 24, 562–577. [Google Scholar] [CrossRef] [PubMed]
- Riedl, S.; Rinner, B.; Schaider, H.; Lohner, K.; Zweytick, D. Killing of melanoma cells and their metastases by human lactoferricin derivatives requires interaction with the cancer marker phosphatidylserine. BioMetals 2014, 27, 981–997. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rascón-Cruz, Q.; Espinoza-Sánchez, E.A.; Siqueiros-Cendón, T.S.; Nakamura-Bencomo, S.I.; Arévalo-Gallegos, S.; Iglesias-Figueroa, B.F. Lactoferrin: A glycoprotein involved in immunomodulation, anticancer, and antimicrobial processes. Molecules 2021, 26, 205. [Google Scholar] [CrossRef]
- Pata, S.; Yaraksa, N.; Daduang, S.; Temsiripong, Y.; Svasti, J.; Araki, T.; Thammasirirak, S. Characterization of the novel antibacterial peptide Leucrocin from crocodile (Crocodylus siamensis) white blood cell extracts. Dev. Comp. Immunol. 2011, 35, 545–553. [Google Scholar] [CrossRef]
- Theansungnoen, T.; Maijaroen, S.; Jangpromma, N.; Yaraksa, N.; Daduang, S.; Temsiripong, T.; Daduang, J.; Klaynongsruang, S. Cationic Antimicrobial Peptides Derived from Crocodylus siamensis Leukocyte Extract, Revealing Anticancer Activity and Apoptotic Induction on Human Cervical Cancer Cells. Protein J. 2016, 35, 202–211. [Google Scholar] [CrossRef]
- Patathananone, S.; Thammasirirak, S.; Daduang, J.; Chung, J.G.; Temsiripong, Y.; Daduang, S. Bioactive compounds from crocodile ( Crocodylus siamensis ) white blood cells induced apoptotic cell death in hela cells. Environ. Toxicol. 2016, 31, 986–997. [Google Scholar] [CrossRef]
- Maijaroen, S.; Jangpromma, N.; Daduang, J.; Klaynongsruang, S. KT2 and RT2 modified antimicrobial peptides derived from Crocodylus siamensis Leucrocin I show activity against human colon cancer HCT-116 cells. Environ. Toxicol. Pharmacol. 2018, 62, 164–176. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Good, D.; Mosaiab, T.; Liu, W.; Ni, G.; Kaur, J.; Liu, X.; Jessop, C.; Yang, L.; Fadhil, R.; et al. Significance of LL-37 on Immunomodulation and Disease Outcome. Biomed Res. Int. 2020, 2020, 8349712. [Google Scholar] [CrossRef]
- Wang, G.; Narayana, J.L.; Mishra, B.; Zhang, Y.; Wang, F.; Wang, C.; Zarena, D.; Lushnikova, T.; Wang, X. Design of Antimicrobial Peptides: Progress Made with Human Cathelicidin LL-37. In Antimicrobial Peptides. Advances in Experimental Medicine and Biology; Matsuzaki, K., Ed.; Springer: Singapore, 2019; Volume 1117, pp. 215–240. [Google Scholar]
- Kuroda, K.; Okumura, K.; Isogai, H.; Isogai, E. The Human Cathelicidin Antimicrobial Peptide LL-37 and Mimics are Potential Anticancer Drugs. Front. Oncol. 2015, 5, 144. [Google Scholar] [CrossRef] [Green Version]
- Hsu, W.H.; Lai, Y.J.; Wu, S.C. Effects of the anti-microbial peptide pardaxin plus sodium erythorbate dissolved in different gels on the quality of Pacific white shrimp under refrigerated storage. Food Control 2017, 73, 712–719. [Google Scholar] [CrossRef]
- Bhunia, A.; Domadia, P.N.; Torres, J.; Hallock, K.J.; Ramamoorthy, A.; Bhattacharjya, S. NMR structure of pardaxin, a pore-forming antimicrobial peptide, in lipopolysaccharide micelles: Mechanism of outer membrane permeabilization. J. Biol. Chem. 2010, 285, 3883–3895. [Google Scholar] [CrossRef] [Green Version]
- Epand, R.F.; Ramamoorthy, A.; Epand, R.M. Membrane Lipid Composition and the Interaction of Pardaxin: The Role of Cholesterol. Protein Pept. Lett. 2005, 13, 1–5. [Google Scholar] [CrossRef]
- Ting, C.H.; Huang, H.N.; Huang, T.C.; Wu, C.J.; Chen, J.Y. The mechanisms by which pardaxin, a natural cationic antimicrobial peptide, targets the endoplasmic reticulum and induces c-FOS. Biomaterials 2014, 35, 3627–3640. [Google Scholar] [CrossRef]
- Wu, S.-P.; Huang, T.-C.; Lin, C.-C.; Hui, C.-F.; Lin, C.-H.; Chen, J.-Y. Pardaxin, a Fish Antimicrobial Peptide, Exhibits Antitumor Activity toward Murine Fibrosarcoma in vitro and In vivo. Mar. Drugs 2012, 10, 1852–1872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, T.C.; Lee, J.F.; Chen, J.Y. Pardaxin, an antimicrobial peptide, triggers caspase-dependent and ROS-mediated apoptosis in HT-1080 cells. Mar. Drugs 2011, 9, 1995–2009. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhandari, D.; Rafiq, S.; Gat, Y.; Gat, P.; Waghmare, R.; Kumar, V. A Review on Bioactive Peptides: Physiological Functions, Bioavailability and Safety. Int. J. Pept. Res. Ther. 2020, 26. [Google Scholar] [CrossRef]
- Parvy, J.P.; Yu, Y.; Dostalova, A.; Kondo, S.; Kurjan, A.; Bulet, P.; Lemaître, B.; Vidal, M.; Cordero, J.B. The antimicrobial peptide defensin cooperates with tumour necrosis factor to drive tumour cell death in Drosophila. eLife 2019, 8, e45061. [Google Scholar] [CrossRef]
- Avilés-Gaxiola, S.; Gutiérrez-Grijalva, E.P.; León-Felix, J.; Angulo-Escalante, M.A.; Heredia, J.B. Peptides in Colorectal Cancer: Current State of Knowledge. Plant Foods Hum. Nutr. 2020, 75, 467–476. [Google Scholar] [CrossRef] [PubMed]
- Ashaolu, T.J. Health Applications of Soy Protein Hydrolysates. Int. J. Pept. Res. Ther. 2020, 26, 2333–2343. [Google Scholar] [CrossRef]
- Wan, X.; Liu, H.; Sun, Y.; Zhang, J.; Chen, X.; Chen, N. Lunasin: A promising polypeptide for the prevention and treatment of cancer. Oncol. Lett. 2017, 13, 3997–4001. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galvez, A.F.; Chen, N.; Macasieb, J.; De Lumen, B.O. Chemopreventive property of a soybean peptide (Lunasin) that binds to deacetylated histones and inhibits acetylation. Cancer Res. 2001, 61, 7473–7478. [Google Scholar] [PubMed]
- Marcela, G.-M. Bioactive Peptides from Legumes as Anticancer Therapeutic Agents. Int. J. Cancer Clin. Res. 2017, 4, 81. [Google Scholar] [CrossRef]
- Rusdi, N.K.; Purwaningsih, E.H.; Hestiantoro, A.; Elya, B.; Kusmardi, K. In vivo antimammary tumor effects of soybean extract with targeted lunasin (ET-Lun). Pharmacogn. J. 2021, 13, 1269–1276. [Google Scholar] [CrossRef]
- Khalifa, S.A.M.; Elias, N.; Farag, M.A.; Chen, L.; Saeed, A.; Hegazy, M.E.F.; Moustafa, M.S.; El-Wahed, A.A.; Al-Mousawi, S.M.; Musharraf, S.G.; et al. Marine natural products: A source of novel anticancer drugs. Mar. Drugs 2019, 17, 491. [Google Scholar] [CrossRef] [Green Version]
- Görgüç, A.; Gençdağ, E.; Yılmaz, F.M. Bioactive peptides derived from plant origin by-products: Biological activities and techno-functional utilizations in food developments—A review. Food Res. Int. 2020, 136, 109504. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Feng, J.; Du, Z.; Zhen, H.; Lin, M.; Jia, S.; Li, T.; Huang, X.; Ostenson, C.G.; Chen, Z. Oral administration of soybean peptide Vglycin normalizes fasting glucose and restores impaired pancreatic function in Type 2 diabetic Wistar rats. J. Nutr. Biochem. 2014, 25, 954–963. [Google Scholar] [CrossRef]
- Gao, C.; Sun, R.; Xie, Y.R.; Jiang, A.L.; Lin, M.; Li, M.; Chen, Z.W.; Zhang, P.; Jin, H.; Feng, J.P. The soy-derived peptide Vglycin inhibits the growth of colon cancer cells in vitro and in vivo. Exp. Biol. Med. 2017, 242, 1034–1043. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abbasi Gharibkandi, N.; Conlon, J.M.; Hosseinimehr, S.J. Strategies for improving stability and pharmacokinetic characteristics of radiolabeled peptides for imaging and therapy. Peptides 2020, 133, 170385. [Google Scholar] [CrossRef]
- Sathya, R.; Mubarakali, D.; Mohamedsaalis, J.; Kim, J.W. A systemic review on microalgal peptides: Bioprocess and sustainable applications. Sustainability 2021, 13, 3262. [Google Scholar] [CrossRef]
- Luong, H.X.; Thanh, T.T.; Tran, T.H. Antimicrobial peptides—Advances in development of therapeutic applications. Life Sci. 2020, 260, 118407. [Google Scholar] [CrossRef]
- Kumar, P.; Kizhakkedathu, J.N.; Straus, S.K. Antimicrobial peptides: Diversity, mechanism of action and strategies to improve the activity and biocompatibility in vivo. Biomolecules 2018, 8, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mathur, D.; Singh, S.; Mehta, A.; Agrawal, P.; Raghava, G.P.S. In silico approaches for predicting the half-life of natural and modified peptides in blood. PLoS ONE 2018, 13, e0196829. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pilleron, S.; Sarfati, D.; Janssen-Heijnen, M.; Vignat, J.; Ferlay, J.; Bray, F.; Soerjomataram, I. Global cancer incidence in older adults, 2012 and 2035: A population-based study. Int. J. Cancer 2019, 144, 49–58. [Google Scholar] [CrossRef]
- Tripathi, P.P.; Arami, H.; Banga, I.; Gupta, J.; Gandhi, S. Cell penetrating peptides in preclinical and clinical cancer diagnosis and therapy. Oncotarget 2018, 9, 37252. [Google Scholar] [CrossRef] [Green Version]
- Zafar, S.; Beg, S.; Panda, S.K.; Rahman, M.; Alharbi, K.S.; Jain, G.K.; Ahmad, F.J. Novel therapeutic interventions in cancer treatment using protein and peptide-based targeted smart systems. Semin. Cancer Biol. 2021, 69, 249–267. [Google Scholar] [CrossRef] [PubMed]
- Sang, M.; Zhang, J.; Zhuge, Q. Selective cytotoxicity of the antibacterial peptide ABP-dHC-Cecropin A and its analog towards leukemia cells. Eur. J. Pharmacol. 2017, 803, 138–147. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.C.; Lim, Y.F.; Russo, E.; Schneider, P.; Bolliger, L.; Edenharter, A.; Altmann, K.H.; Halin, C.; Hiss, J.A.; Schneider, G. Multidimensional Design of Anticancer Peptides. Angew. Chem. Int. Ed. 2015, 35, 10370–10374. [Google Scholar] [CrossRef] [PubMed]
- Trinidad-Calderón, P.A.; Acosta-Cruz, E.; Rivero-Masante, M.N.; Díaz-Gómez, J.L.; García-Lara, S.; López-Castillo, L.M. Maize bioactive peptides: From structure to human health. J. Cereal Sci. 2021, 100, 103232. [Google Scholar] [CrossRef]
- Robey, R.W.; Pluchino, K.M.; Hall, M.D.; Fojo, A.T.; Bates, S.E.; Gottesman, M.M. Revisiting the role of ABC transporters in multidrug-resistant cancer. Nat. Rev. Cancer 2018, 18, 452–464. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Gohain, N.; Chen, S.; Li, Y.; Zhao, X.; Li, B.; Tolbert, W.D.; He, W.; Pazgier, M.; Hu, H.; et al. Design of ultrahigh-affinity and dual-specificity peptide antagonists of MDM2 and MDMX for P53 activation and tumor suppression. Acta Pharm. Sin. B 2021, 11, 2655–2669. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Qin, X.; Yang, D.; Jiang, Y.; Zheng, W.; Wang, D.; Tian, Y.; Liu, Q.; Xu, N.; Li, Z. The development of activatable lytic peptides for targeting triple negative breast cancer. Cell Death Discov. 2017, 3, 17037. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khine, H.E.E.; Ecoy, G.A.U.; Roytrakul, S.; Phaonakrop, N.; Pornputtapong, N.; Prompetchara, E.; Chanvorachote, P.; Chaotham, C. Chemosensitizing activity of peptide from Lentinus squarrosulus (Mont.) on cisplatin-induced apoptosis in human lung cancer cells. Sci. Rep. 2021, 11, 4060. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Trinidad-Calderón, P.A.; Varela-Chinchilla, C.D.; García-Lara, S. Natural Peptides Inducing Cancer Cell Death: Mechanisms and Properties of Specific Candidates for Cancer Therapeutics. Molecules 2021, 26, 7453. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules26247453
Trinidad-Calderón PA, Varela-Chinchilla CD, García-Lara S. Natural Peptides Inducing Cancer Cell Death: Mechanisms and Properties of Specific Candidates for Cancer Therapeutics. Molecules. 2021; 26(24):7453. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules26247453
Chicago/Turabian StyleTrinidad-Calderón, Plinio A., Carlos Daniel Varela-Chinchilla, and Silverio García-Lara. 2021. "Natural Peptides Inducing Cancer Cell Death: Mechanisms and Properties of Specific Candidates for Cancer Therapeutics" Molecules 26, no. 24: 7453. https://0-doi-org.brum.beds.ac.uk/10.3390/molecules26247453