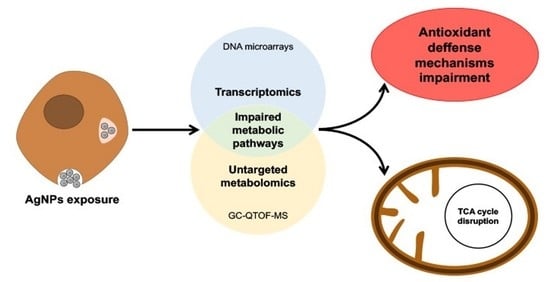
A Multi-Omics Approach to Evaluate the Toxicity Mechanisms Associated with Silver Nanoparticles Exposure
Abstract
:1. Introduction
2. Materials and Methods
2.1. Nanoparticles: Physicochemical Characterization and Stability
2.2. Cell Culture and Treatment
2.3. Cytotoxicity Assays
2.4. Targeted Metabolomics
2.5. Untargeted Metabolomics
2.6. Transcriptome Analysis
3. Results
3.1. Physicochemical Characterization and Stability of the AgNPs
3.2. Cytotoxicity of AgNPs
3.3. Targeted Metabolomics
3.4. Untargeted Metabolomics Results
3.5. Transcriptome Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Zhang, X.; Ge, M.; Dong, J.; Huang, J.; He, J.-H.; Lai, Y. Polydopamine-Inspired Design and Synthesis of Visible-Light-Driven Ag NPs@C@elongated TiO2 NTs Core–Shell Nanocomposites for Sustainable Hydrogen Generation. ACS Sustain. Chem. Eng. 2019, 7, 558–568. [Google Scholar] [CrossRef]
- Ge, M.-Z.; Cao, C.-Y.; Li, S.-H.; Tang, Y.-X.; Wang, L.-N.; Qi, N.; Huang, J.-Y.; Zhang, K.-Q.; Al-Deyab, S.S.; Lai, Y.-K. In situ plasmonic Ag nanoparticle anchored TiO2 nanotube arrays as visible-light-driven photocatalysts for enhanced water splitting. Nanoscale 2016, 8, 5226–5234. [Google Scholar] [CrossRef] [PubMed]
- Ge, M.; Cao, C.; Huang, J.; Zhang, X.; Tang, Y.; Zhou, X.; Zhang, K.; Chen, Z.; Lai, Y. Rational design of materials interface at nanoscale towards intelligent oil–water separation. Nanoscale Horiz. 2018, 3, 235–260. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-F.; Liu, Z.-G.; Shen, W.; Gurunathan, S. Silver Nanoparticles: Synthesis, Characterization, Properties, Applications, and Therapeutic Approaches. Int. J. Mol. Sci. 2016, 17, 1534. [Google Scholar] [CrossRef]
- Lee, S.H.; Jun, B.-H. Silver Nanoparticles: Synthesis and Application for Nanomedicine. Int. J. Mol. Sci. 2019, 20, 865. [Google Scholar] [CrossRef] [Green Version]
- Abbasi, E.; Milani, M.; Aval, S.F.; Kouhi, M.; Akbarzadeh, A.; Nasrabadi, H.T.; Nikasa, P.; Joo, S.W.; Hanifehpour, Y.; Nejati-Koshki, K.; et al. Silver nanoparticles: Synthesis methods, bio-applications and properties. Crit. Rev. Microbiol. 2016, 42, 173–180. [Google Scholar] [CrossRef]
- Ferdous, Z.; Nemmar, A. Health Impact of Silver Nanoparticles: A Review of the Biodistribution and Toxicity Following Various Routes of Exposure. Int. J. Mol. Sci. 2020, 21, 2375. [Google Scholar] [CrossRef] [Green Version]
- Mathur, P.; Jha, S.; Ramteke, S.; Jain, N.K. Pharmaceutical aspects of silver nanoparticles. Artif. Cells Nanomed. Biotechnol. 2018, 46, 115–126. [Google Scholar] [CrossRef] [Green Version]
- De Matteis, V.; Malvindi, M.A.; Galeone, A.; Brunetti, V.; De Luca, E.; Kote, S.; Kshirsagar, P.; Sabella, S.; Bardi, G.; Pompa, P.P. Negligible particle-specific toxicity mechanism of silver nanoparticles: The role of Ag+ ion release in the cytosol. Nanomed. Nanotechnol. Biol. Med. 2015, 11, 731–739. [Google Scholar] [CrossRef]
- Xue, Y.; Zhang, T.; Zhang, B.; Gong, F.; Huang, Y.; Tang, M. Cytotoxicity and apoptosis induced by silver nanoparticles in human liver HepG2 cells in different dispersion media. J. Appl. Toxicol. 2016, 36, 352–360. [Google Scholar] [CrossRef]
- Bastos, V.; Duarte, I.; Santos, C.; Oliveira, H. A study of the effects of citrate-coated silver nanoparticles on RAW 264.7 cells using a toolbox of cytotoxic endpoints. J. Nanoparticle Res. 2017, 19, 163. [Google Scholar] [CrossRef]
- Fernández, M.N.; Muñoz-Olivas, R.; Luque-Garcia, J.L. SILAC-based quantitative proteomics identifies size-dependent molecular mechanisms involved in silver nanoparticles-induced toxicity. Nanotoxicology 2019, 13, 812–826. [Google Scholar] [CrossRef] [PubMed]
- Viant, M.R.; Ebbels, T.M.D.; Beger, R.D.; Ekman, D.R.; Epps, D.J.T.; Kamp, H.; Leonards, P.E.G.; Loizou, G.D.; Macrae, J.I.; Van Ravenzwaay, B.; et al. Use cases, best practice and reporting standards for metabolomics in regulatory toxicology. Nat. Commun. 2019, 10, 3041. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Li, C.; Yao, C.; Ding, L.; Lei, Z.; Wu, M.; Yanli, W.; Chenchen, L.; ChenJie, Y.; Lin, D.; et al. Techniques for Investigating Molecular Toxicology of Nanomaterials. J. Biomed. Nanotechnol. 2016, 12, 1115–1135. [Google Scholar] [CrossRef]
- Robertson, D.G.; Watkins, P.B.; Reily, M.D. Metabolomics in Toxicology: Preclinical and Clinical Applications. Toxicol. Sci. 2011, 120, S146–S170. [Google Scholar] [CrossRef] [Green Version]
- Schrimpe-Rutledge, A.C.; Codreanu, S.G.; Sherrod, S.D.; McLean, J.A. Untargeted Metabolomics Strategies—Challenges and Emerging Directions. J. Am. Soc. Mass Spectrom. 2016, 27, 1897–1905. [Google Scholar] [CrossRef] [Green Version]
- Joseph, P. Transcriptomics in toxicology. Food Chem. Toxicol. 2017, 109, 650–662. [Google Scholar] [CrossRef]
- Cui, Y.; Paules, R.S. Use of transcriptomics in understanding mechanisms of drug-induced toxicity. Pharmacogenomics 2010, 11, 573–585. [Google Scholar] [CrossRef] [Green Version]
- Estevez, H.; Garcia-Calvo, E.; Rivera-Torres, J.; Vallet-Regí, M.; González, B.; Luque-Garcia, J. Transcriptome Analysis Identifies Novel Mechanisms Associated with the Antitumor Effect of Chitosan-Stabilized Selenium Nanoparticles. Pharmaceutics 2021, 13, 356. [Google Scholar] [CrossRef]
- Machuca, A.; Garcia-Calvo, E.; Anunciação, D.S.; Luque-Garcia, J.L. Integration of Transcriptomics and Metabolomics to Reveal the Molecular Mechanisms Underlying Rhodium Nanoparticles-Based Photodynamic Cancer Therapy. Pharmaceutics 2021, 13, 1629. [Google Scholar] [CrossRef]
- Carrola, J.; Bastos, V.; Jarak, I.; Oliveira-Silva, R.; Malheiro, E.; Daniel-Da-Silva, A.L.; Oliveira, H.; Santos, C.; Gil, A.M.; Duarte, I.F.; et al. Metabolomics of silver nanoparticles toxicity in HaCaT cells: Structure–activity relationships and role of ionic silver and oxidative stress. Nanotoxicology 2016, 10, 1105–1117. [Google Scholar] [CrossRef] [PubMed]
- Foldbjerg, R.; Irving, E.S.; Hayashi, Y.; Sutherland, D.S.; Thorsen, K.; Autrup, H.; Beer, C. Global Gene Expression Profiling of Human Lung Epithelial Cells After Exposure to Nanosilver. Toxicol. Sci. 2012, 130, 145–157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia-Calvo, E.; Machuca, A.; Nerín, C.; Rosales-Conrado, N.; Anunciação, D.S.; Luque-Garcia, J.L. Integration of untargeted and targeted mass spectrometry-based metabolomics provides novel insights into the potential toxicity associated to surfynol. Food Chem. Toxicol. 2020, 146, 111849. [Google Scholar] [CrossRef] [PubMed]
- Sapcariu, S.C.; Kanashova, T.; Weindl, D.; Ghelfi, J.; Dittmar, G.; Hiller, K. Simultaneous extraction of proteins and metabolites from cells in culture. MethodsX 2014, 1, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Larson, R.; Farber, B. Elementary Statistics: Picturing the World, 6th ed.; Pearson: London, UK, 2015; pp. 489–541. [Google Scholar]
- Suliman, Y.A.O.; Ali, D.; Alarifi, S.; Harrath, A.H.; Mansour, L.; Alwasel, S.H. Evaluation of cytotoxic, oxidative stress, proinflammatory and genotoxic effect of silver nanoparticles in human lung epithelial cells. Environ. Toxicol. 2013, 30, 149–160. [Google Scholar] [CrossRef]
- Daher, B.; Vučetić, M.; Pouysségur, J. Cysteine Depletion, a Key Action to Challenge Cancer Cells to Ferroptotic Cell Death. Front. Oncol. 2020, 10, 723. [Google Scholar] [CrossRef]
- Novera, W.; Lee, Z.-W.; Nin, D.S.; Dai, M.Z.-Y.; Idres, S.B.; Wu, H.; Damen, J.M.A.; Tan, T.Z.; Sim, A.Y.L.; Long, Y.C.; et al. Cysteine Deprivation Targets Ovarian Clear Cell Carcinoma Via Oxidative Stress and Iron−Sulfur Cluster Biogenesis Deficit. Antioxid. Redox Signal. 2020, 33, 1191–1208. [Google Scholar] [CrossRef]
- Wallis, K.F.; Morehead, L.C.; Bird, J.T.; Byrum, S.D.; Miousse, I.R. Differences in cell death in methionine versus cysteine depletion. Environ. Mol. Mutagen. 2021, 62, 216–226. [Google Scholar] [CrossRef]
- Niu, W.; Chen, F.; Wang, J.; Qian, J.; Yan, S. Antitumor effect of sikokianin C, a selective cystathionine β-synthase inhibitor, against human colon cancer in vitro and in vivo. MedChemComm 2018, 9, 113–120. [Google Scholar] [CrossRef]
- Anderton, B.; Camarda, R.; Balakrishnan, S.; Balakrishnan, A.; Kohnz, R.A.; Lim, L.; Evason, K.J.; Momcilovic, O.; Kruttwig, K.; Huang, Q.; et al. MYC -driven inhibition of the glutamate-cysteine ligase promotes glutathione depletion in liver cancer. EMBO Rep. 2017, 18, 569–585. [Google Scholar] [CrossRef]
- Sun, J.; Zhou, C.; Ma, Q.; Chen, W.; Atyah, M.; Yin, Y.; Fu, P.; Liu, S.; Hu, B.; Ren, N.; et al. High GCLC level in tumor tissues is associated with poor prognosis of hepatocellular carcinoma after curative resection. J. Cancer 2019, 10, 3333–3343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Quan, C.; Wang, C.; Wu, H.; Li, Z.; Ye, Q. Systematic manipulation of glutathione metabolism in Escherichia coli for improved glutathione production. Microb. Cell Fact. 2016, 15, 38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sikalidis, A.; Mazor, K.M.; Lee, J.-I.; Roman, H.B.; Hirschberger, L.L.; Stipanuk, M.H. Upregulation of capacity for glutathione synthesis in response to amino acid deprivation: Regulation of glutamate-cysteine ligase subunits. Amino Acids 2014, 46, 1285–1296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gwangwa, M.V.; Joubert, A.M.; Visagie, M.H. Effects of glutamine deprivation on oxidative stress and cell survival in breast cell lines. Biol. Res. 2019, 52, 15. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Alesi, G.N.; Kang, S. Glutaminolysis as a target for cancer therapy. Oncogene 2016, 35, 3619–3625. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitra, D.; Vega-Rubin-De-Celis, S.; Royla, N.; Bernhardt, S.; Wilhelm, H.; Tarade, N.; Poschet, G.; Buettner, M.; Binenbaum, I.; Borgoni, S.; et al. Abrogating GPT2 in triple-negative breast cancer inhibits tumor growth and promotes autophagy. Int. J. Cancer 2021, 148, 1993–2009. [Google Scholar] [CrossRef] [PubMed]
- Fan, S.; Wang, Y.; Zhang, Z.; Lu, J.; Wu, Z.; Shan, Q.; Sun, C.; Wu, D.; Li, M.; Sheng, N.; et al. High expression of glutamate-ammonia ligase is associated with unfavorable prognosis in patients with ovarian cancer. J. Cell. Biochem. 2018, 119, 6008–6015. [Google Scholar] [CrossRef]
- Tyrakis, P.A.; Yurkovich, M.E.; Sciacovelli, M.; Papachristou, E.K.; Bridges, H.R.; Gaude, E.; Schreiner, A.; D’Santos, C.; Hirst, J.; Hernandez-Fernaud, J.; et al. Fumarate Hydratase Loss Causes Combined Respiratory Chain Defects. Cell Rep. 2017, 21, 1036–1047. [Google Scholar] [CrossRef] [Green Version]
- Bergaggio, E.; Riganti, C.; Garaffo, G.; Vitale, N.; Mereu, E.; Bandini, C.; Pellegrino, E.; Pullano, V.; Omedè, P.; Todoerti, K.; et al. IDH2 inhibition enhances proteasome inhibitor responsiveness in hematological malignancies. Blood 2019, 133, 156–167. [Google Scholar] [CrossRef] [Green Version]
- Ku, H.J.; Kwon, O.-S.; Kang, B.S.; Lee, D.S.; Lee, H.-S.; Park, J.-W. IDH2 knockdown sensitizes tumor cells to emodin cytotoxicity in vitro and in vivo. Free Radic. Res. 2016, 50, 1089–1097. [Google Scholar] [CrossRef]
- Gunter, E.T.; Gavin, C.E.; Gunter, K.K. The role of mitochondrial oxidative stress and ATP depletion in the pathology of manganese toxicity. In Metal Ion in Stroke; Li, Y., Zhang, J., Eds.; Springer: New York, NY, USA, 2012; pp. 591–606. [Google Scholar]
- Del Nagro, C.; Xiao, Y.; Rangell, L.; Reichelt, M.; O’Brien, T. Depletion of the Central Metabolite NAD Leads to Oncosis-mediated Cell Death. J. Biol. Chem. 2014, 289, 35182–35192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia-Bermudez, J.; Baudrier, L.; La, K.; Zhu, X.G.; Fidelin, J.; Sviderskiy, V.O.; Papagiannakopoulos, T.; Molina, H.; Snuderl, M.; Lewis, C.A.; et al. Aspartate is a limiting metabolite for cancer cell proliferation under hypoxia and in tumours. Nat. Cell Biol. 2018, 20, 775–781. [Google Scholar] [CrossRef] [PubMed]
- Alkan, H.F.; Walter, K.E.; Luengo, A.; Madreiter-Sokolowski, C.T.; Stryeck, S.; Lau, A.N.; Al-Zoughbi, W.; Lewis, C.A.; Thomas, C.J.; Hoefler, G.; et al. Cytosolic Aspartate Availability Determines Cell Survival When Glutamine Is Limiting. Cell Metab. 2018, 28, 706–720. [Google Scholar] [CrossRef] [Green Version]
- Cheng, C.-T.; Qi, Y.; Wang, Y.-C.; Chi, K.; Chung, Y.; Ouyang, C.; Chen, Y.-R.; Oh, M.E.; Sheng, X.; Tang, Y.; et al. Arginine starvation kills tumor cells through aspartate exhaustion and mitochondrial dysfunction. Commun. Biol. 2018, 1, 178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delage, B.; Fennell, D.A.; Nicholson, L.; McNeish, I.; Lemoine, N.R.; Crook, T.; Szlosarek, P.W. Arginine deprivation and argininosuccinate synthetase expression in the treatment of cancer. Int. J. Cancer 2010, 126, 2762–2772. [Google Scholar] [CrossRef] [PubMed]
- Burke, L.; Guterman, I.; Gallego, R.P.; Britton, R.G.; Burschowsky, D.; Tufarelli, C.; Rufini, A. The Janus-like role of proline metabolism in cancer. Cell Death Discov. 2020, 6, 104. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Zuo, W.; Wang, X.; Zhang, Q.; Gan, X.; Tan, N.; Jia, W.; Liu, J.; Li, Z.; Zhou, B.; et al. Deciphering the effects of PYCR1 on cell function and its associated mechanism in hepatocellular carcinoma. Int. J. Biol. Sci. 2021, 17, 2223–2239. [Google Scholar] [CrossRef]
- Zhang, Q.; Yin, Y.; Zhao, H.; Shi, Y.; Zhang, W.; Yang, Z.; Liu, T.; Huang, Y.; Yu, Z. P4HA1 regulates human colorectal cancer cells through HIF1α-mediated Wnt signaling. Oncol. Lett. 2021, 21, 145. [Google Scholar] [CrossRef]
- Agarwal, S.; Behring, M.; Kim, H.-G.; Bajpai, P.; Chakravarthi, B.V.; Gupta, N.; Elkholy, A.; Al Diffalha, S.; Varambally, S.; Manne, U. Targeting P4HA1 with a Small Molecule Inhibitor in a Colorectal Cancer PDX Model. Transl. Oncol. 2020, 13, 100754. [Google Scholar] [CrossRef]
- Sahu, N.; Cruz, D.D.; Gao, M.; Sandoval, W.; Haverty, P.M.; Liu, J.; Stephan, J.-P.; Haley, B.; Classon, M.; Hatzivassiliou, G.; et al. Proline Starvation Induces Unresolved ER Stress and Hinders mTORC1-Dependent Tumorigenesis. Cell Metab. 2016, 24, 753–761. [Google Scholar] [CrossRef] [Green Version]
- Xiao, F.; Wang, C.; Yin, H.; Yu, J.; Chen, S.; Fang, J.; Guo, F. Leucine deprivation inhibits proliferation and induces apoptosis of human breast cancer cells via fatty acid synthase. Oncotarget 2016, 7, 63679–63689. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kobayashi, D.; Kusama, M.; Onda, M.; Nakahata, N. The Effect of Pantothenic Acid Deficiency on Keratinocyte Proliferation and the Synthesis of Keratinocyte Growth Factor and Collagen in Fibroblasts. J. Pharmacol. Sci. 2011, 115, 230–234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.-J.; Yu, G.; Jiang, L.; Li, T.; Lin, Q.; Tang, Y.; Gu, W. p53-dependent regulation of metabolic function through transcriptional activation of pantothenate kinase-1 gene. Cell Cycle 2013, 12, 753–761. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Graham, N.A.; Tahmasian, M.; Kohli, B.; Komisopoulou, E.; Zhu, M.; Vivanco, I.; Teitell, M.A.; Wu, H.; Ribas, A.; Lo, R.S.; et al. Glucose deprivation activates a metabolic and signaling amplification loop leading to cell death. Mol. Syst. Biol. 2012, 8, 589. [Google Scholar] [CrossRef] [PubMed]
- Dai, W.; Xu, Y.; Mo, S.; Li, Q.; Yu, J.; Wang, R.; Ma, Y.; Ni, Y.; Xiang, W.; Han, L.; et al. GLUT3 induced by AMPK/CREB1 axis is key for withstanding energy stress and augments the efficacy of current colorectal cancer therapies. Signal. Transduct. Target. Ther. 2020, 5, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Low, B.S.J.; Lim, C.S.; Ding, S.S.L.; Tan, Y.S.; Ng, N.H.J.; Krishnan, V.G.; Ang, S.F.; Neo, C.W.Y.; Verma, C.S.; Hoon, S.; et al. Decreased GLUT2 and glucose uptake contribute to insulin secretion defects in MODY3/HNF1A hiPSC-derived mutant β cells. Nat. Commun. 2021, 12, 177. [Google Scholar] [CrossRef] [PubMed]
- Kuang, R.; Jahangiri, A.; Mascharak, S.; Nguyen, A.; Chandra, A.; Flanigan, P.M.; Yagnik, G.; Wagner, J.R.; De Lay, M.; Carrera, D.; et al. GLUT3 upregulation promotes metabolic reprogramming associated with antiangiogenic therapy resistance. JCI Insight 2017, 2, e88815. [Google Scholar] [CrossRef]
- DeWaal, D.; Nogueira, V.; Terry, A.R.; Patra, K.C.; Jeon, S.M.; Guzman, G.; Au, J.; Long, C.P.; Antoniewicz, M.R.; Hay, N. Hexokinase-2 depletion inhibits glycolysis and induces oxidative phosphorylation in hepatocellular carcinoma and sensitizes to metformin. Nat. Commun. 2018, 9, 446. [Google Scholar] [CrossRef]
- Feng, Y.; Zhang, Y.; Cai, Y.; Liu, R.; Lu, M.; Li, T.; Fu, Y.; Guo, M.; Huang, H.; Ou, Y.; et al. A20 targets PFKL and glycolysis to inhibit the progression of hepatocellular carcinoma. Cell Death Dis. 2020, 11, 89. [Google Scholar] [CrossRef]
- Bartrons, R.; Caro, J. Hypoxia, glucose metabolism and the Warburg’s effect. J. Bioenerg. Biomembr. 2007, 39, 223–229. [Google Scholar] [CrossRef]
- Yi, M.; Ban, Y.; Tan, Y.; Xiong, W.; Li, G.; Xiang, B. 6-Phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 and 4: A pair of valves for fine-tuning of glucose metabolism in human cancer. Mol. Metab. 2019, 20, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Fan, K.; Wang, J.; Sun, W.; Shen, S.; Ni, X.; Gong, Z.; Zheng, B.; Gao, Z.; Ni, X.; Suo, T.; et al. MUC16 C-terminal binding with ALDOC disrupts the ability of ALDOC to sense glucose and promotes gallbladder carcinoma growth. Exp. Cell Res. 2020, 394, 112118. [Google Scholar] [CrossRef] [PubMed]
- Grüning, N.-M.; Du, D.; Keller, M.A.; Luisi, B.F.; Ralser, M. Inhibition of triosephosphate isomerase by phosphoenolpyruvate in the feedback-regulation of glycolysis. Open Biol. 2014, 4, 130232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, Y.; Wu, C.; Yang, J.; Zhao, Y.; Jia, H.; Xue, M.; Xu, D.; Yang, F.; Fu, D.; Wang, C.; et al. Insulin-like growth factor 1-induced enolase 2 deacetylation by HDAC3 promotes metastasis of pancreatic cancer. Signal. Transduct. Target. Ther. 2020, 5, 53. [Google Scholar] [CrossRef]
- Nguyen, A.; Loo, J.M.; Mital, R.; Weinberg, E.M.; Man, F.Y.; Zeng, Z.; Paty, P.B.; Saltz, L.; Janjigian, Y.Y.; De Stanchina, E.; et al. PKLR promotes colorectal cancer liver colonization through induction of glutathione synthesis. J. Clin. Investig. 2016, 126, 681–694. [Google Scholar] [CrossRef] [Green Version]
- Guo, T.; Chen, T.; Gu, C.; Li, B.; Xu, C. Genetic and molecular analyses reveal G6PC as a key element connecting glucose metabolism and cell cycle control in ovarian cancer. Tumor Biol. 2015, 36, 7649–7658. [Google Scholar] [CrossRef]
- Jin, G.-Z.; Zhang, Y.; Cong, W.-M.; Wu, X.; Wang, X.; Wu, S.; Wang, S.; Zhou, W.; Yuan, S.; Gao, H.; et al. Phosphoglucomutase 1 inhibits hepatocellular carcinoma progression by regulating glucose trafficking. PLoS Biol. 2018, 16, e2006483. [Google Scholar] [CrossRef] [Green Version]
- Lee, C.-H.; Jeong, S.-J.; Yun, S.-M.; Kim, J.-H.; Lee, H.-J.; Ahn, K.S.; Won, S.-H.; Kim, H.S.; Ahn, K.-S.; Zhu, S.; et al. Down-regulation of phosphoglucomutase 3 mediates sulforaphane-induced cell death in LNCaP prostate cancer cells. Proteome Sci. 2010, 8, 67. [Google Scholar] [CrossRef] [Green Version]
- Ricciardiello, F.; Gang, Y.; Palorini, R.; Li, Q.; Giampà, M.; Zhao, F.; You, L.; La Ferla, B.; De Vitto, H.; Guan, W.; et al. Hexosamine pathway inhibition overcomes pancreatic cancer resistance to gemcitabine through unfolded protein response and EGFR-Akt pathway modulation. Oncogene 2020, 39, 4103–4117. [Google Scholar] [CrossRef]
- Zhang, K.; Liu, L.; Wang, M.; Yang, M.; Li, X.; Xia, X.; Tian, J.; Tan, S.; Luo, L. A novel function of IMPA2, plays a tumor-promoting role in cervical cancer. Cell Death Dis. 2020, 11, 371. [Google Scholar] [CrossRef]
- Sharma, I.; Deng, F.; Liao, Y.; Kanwar, Y.S. Myo-inositol Oxygenase (MIOX) Overexpression Drives the Progression of Renal Tubulointerstitial Injury in Diabetes. Diabetes 2020, 69, 1248–1263. [Google Scholar] [CrossRef] [PubMed]






| Metabolite Name | Retention Time (min) | NIST Rmatch | RM |
|---|---|---|---|
| Glutamine | 19.890 | 874 | 0.69 |
| Threitol | 17.864 | 851 | 0.62 |
| Xylofuranose | 32.638 | 793 | 0.48 |
| L-Cysteine | 18.711 | 868 | 0.33 |
| D-Glucose | 24.964 | 870 | 0.31 |
| L-Threonine | 15.285 | 865 | 0.30 |
| L-Proline | 18.014 | 881 | 0.25 |
| L-Aspartic acid | 18.086 | 791 | 0.24 |
| D-Fructose | 24.294 | 767 | 0.23 |
| β-D-Glucopyranose | 26.375 | 890 | 0.15 |
| D-Galactose | 25.192 | 848 | 0.15 |
| Pantothenic acid | 26.473 | 887 | 0.13 |
| Malic acid | 17.466 | 728 | 0.13 |
| Rythonic acid | 18.541 | 767 | 0.12 |
| Myo-inositol | 27.874 | 830 | 0.10 |
| Octadecanoic acid | 30.225 | 839 | 0.09 |
| Myo-inositol phosphate | 32.426 | 827 | 0.07 |
| L-Tyrosine | 25.543 | 782 | 0.06 |
| Phosphoric acid | 22.634 | 868 | 0.05 |
| L-Leucine | 19.177 | 700 | 0.03 |
| Gene Name | Gene Code | FC |
|---|---|---|
| Phosphofructokinase, platelet | PFKP | 33.45 |
| Hexokinase 1 | HK1 | 11.86 |
| 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 | PFKFB1 | 4.62 |
| Glutamate-cysteine ligase, catalytic subunit | GCLC | 3.49 |
| Enolase 2 (gamma, neuronal) | ENO2 | 2.90 |
| Solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11 | SLC7A11 | 2.66 |
| Glutathione synthetase | GSS | 2.16 |
| Myo-inositol oxygenase | MIOX | 2.08 |
| Solute carrier family 2 (facilitated glucose transporter), member 3 | SLC2A3 | 2.07 |
| Glutaminase | GLS1 | 0.98 |
| Glutaminase 2 (liver, mitochondrial) | GLS2 | 0.92 |
| Cystathionine gamma-lyase | CTH | 0.70 |
| Triosephosphate isomerase 1 | TPI1 | 0.47 |
| Phosphoglucomutase 3 | PGM3 | 0.45 |
| Inositol-3-phosphate synthase 1 | INO1 | 0.43 |
| Pantothenate kinase 3 | PANK3 | 0.42 |
| Fumarate hydratase | FH | 0.41 |
| Isocitrate dehydrogenase 2 (NADP+), mitochondrial | IDH2 | 0.41 |
| Phosphofructokinase, liver | PFKL | 0.40 |
| Cystathionine-beta-synthase | CBS | 0.37 |
| Glutamate-ammonia ligase | GLUL | 0.36 |
| Hexokinase 2 | HK2 | 0.36 |
| Prolyl 4-hydroxylase, alpha polypeptide I | P4HA1 | 0.35 |
| Inositol(myo)-1(or 4)-monophosphatase 2 | IMPA2 | 0.33 |
| Solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 | SLC7A2 | 0.28 |
| Pyrroline-5-carboxylate reductase 1 | PYCR1 | 0.28 |
| Fatty acid synthase | FASN | 0.27 |
| Isocitrate dehydrogenase 1 (NADP+) | IDH1 | 0.23 |
| Solute carrier family 2 (facilitated glucose transporter), member 2 | SLC2A2 | 0.23 |
| Pyruvate kinase, liver and RBC | PKLR | 0.18 |
| Glutamate dehydrogenase 1 | GLUD1 | 0.17 |
| Glutamate dehydrogenase 2 | GLUD2 | 0.17 |
| Phosphoglucomutase 1 | PGM1 | 0.14 |
| Aldolase C, fructose-bisphosphate | ALDOC | 0.13 |
| Argininosuccinate synthase 1 | ASS1 | 0.12 |
| Aldehyde dehydrogenase 18 family, member A1 | ALDH18A1 | 0.12 |
| Glutamic pyruvate transaminase (alanine aminotransferase) 2 | GPT2 | 0.11 |
| Pantothenate kinase 1 | PANK1 | 0.11 |
| 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4; microRNA 6823 | PFKFB4 | 0.10 |
| Glucose-6-phosphatase, catalytic subunit | G6PC | 0.02 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aragoneses-Cazorla, G.; Buendia-Nacarino, M.P.; Mena, M.L.; Luque-Garcia, J.L. A Multi-Omics Approach to Evaluate the Toxicity Mechanisms Associated with Silver Nanoparticles Exposure. Nanomaterials 2022, 12, 1762. https://0-doi-org.brum.beds.ac.uk/10.3390/nano12101762
Aragoneses-Cazorla G, Buendia-Nacarino MP, Mena ML, Luque-Garcia JL. A Multi-Omics Approach to Evaluate the Toxicity Mechanisms Associated with Silver Nanoparticles Exposure. Nanomaterials. 2022; 12(10):1762. https://0-doi-org.brum.beds.ac.uk/10.3390/nano12101762
Chicago/Turabian StyleAragoneses-Cazorla, Guillermo, M. Pilar Buendia-Nacarino, Maria L. Mena, and Jose L. Luque-Garcia. 2022. "A Multi-Omics Approach to Evaluate the Toxicity Mechanisms Associated with Silver Nanoparticles Exposure" Nanomaterials 12, no. 10: 1762. https://0-doi-org.brum.beds.ac.uk/10.3390/nano12101762