Unraveling the Mechanisms of Ch-SeNP Cytotoxicity against Cancer Cells: Insights from Targeted and Untargeted Metabolomics
Abstract
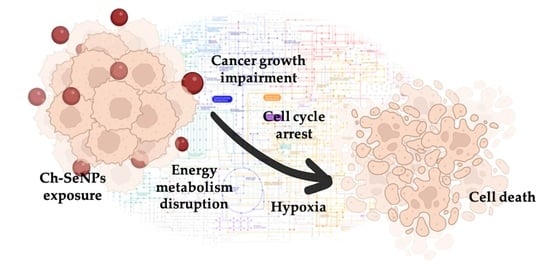
:1. Introduction
2. Materials and Methods
2.1. Synthesis and Characterization of Ch-SeNPs
2.2. Cell-Culture and Exposure Conditions
2.3. Cell Viability
2.4. Targeted Metabolomic Analysis
2.4.1. Extraction of Energy-Related Metabolites
2.4.2. Mass Spectrometry (LC-QqQ-MS) Analysis
2.4.3. Statistical Analysis
2.5. Untargeted Metabolomics
2.5.1. Sample Preparation
2.5.2. High-Resolution Mass spectrometry (GC-TOF-MS) Analysis
2.5.3. Data Treatment and Statistical Analysis
3. Results
3.1. Characterization of Ch-SeNPs
3.2. Cytotoxicity of Ch-SeNPs
3.3. Evaluation of Levels of Energy-Related Metabolites
3.4. Identification of Metabolic Alterations in Cells Exposed to Ch-SeNPs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Johnson, C.H.; Ivanisevic, J.; Siuzdak, G. Metabolomics: Beyond biomarkers and towards mechanisms. Nat. Rev. Mol. Cell Biol. 2016, 17, 451–459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Q.; Vijayakumar, A.; Kahn, B.B. Metabolites as regulators of insulin sensitivity and metabolism. Nat. Rev. Mol. Cell Biol. 2018, 19, 654–672. [Google Scholar] [CrossRef] [PubMed]
- Jacob, F.; Monod, J. Genetic regulatory mechanisms in the synthesis of proteins. JMB 1961, 3, 318–356. [Google Scholar] [CrossRef]
- Rinschen, M.M.; Ivanisevic, J.; Giera, M.; Siuzdak, G. Identification of bioactive metabolites using activity metabolomics. Nat. Rev. Mol. Cell Biol. 2019, 20, 353–367. [Google Scholar] [CrossRef] [PubMed]
- Roberts, L.D.; Souza, A.L.; Gerszten, R.E.; Clish, C.B. Targeted metabolomics. In Current Protocols in Molecular Biology; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2012; Chapter 30, Unit 30.2–30.2.24. [Google Scholar] [CrossRef]
- Di Minno, A.; Gelzo, M.; Stornaiuolo, M.; Ruoppolo, M.; Castaldo, G. The evolving landscape of untargeted metabolomics. NMCD 2021, 31, 1645–1652. [Google Scholar] [CrossRef]
- Schrimpe-Rutledge, A.C.; Codreanu, S.G.; Sherrod, S.D.; McLean, J.A. Untargeted Metabolomics Strategies-Challenges and Emerging Directions. J. Am. Soc. Mass Spectrom. 2016, 27, 1897–1905. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dai, X.; Yu, L.; Zhao, X.; Ostrikov, K.K. Nanomaterials for oncotherapies targeting the hallmarks of cancer. Nanotechnology 2020, 31, 392001. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Tang, Z.; Zhang, D.; Song, W.; Zhang, Y.; Yang, Y.; Ahmad, Z.; Chen, X. Pharmacokinetics, biodistribution and in vivo efficacy of cisplatin loaded poly(L-glutamic acid)-g-methoxy poly(ethylene glycol) complex nanoparticles for tumor therapy. J. Control. Release 2015, 205, 89–97. [Google Scholar] [CrossRef]
- Estevez, H.; Garcia-Lidon, J.C.; Luque-Garcia, J.L.; Camara, C. Effects of chitosan-stabilized selenium nanoparticles on cell proliferation, apoptosis and cell cycle pattern in HepG2 cells: Comparison with other selenospecies. Colloids Surf. B 2014, 122, 184–193. [Google Scholar] [CrossRef]
- Bai, Y.; Wang, Y.; Zhou, Y.; Li, W.; Zheng, W. Modification and modulation of saccharides on elemental selenium nanoparticles in liquid phase. Mater. Lett. 2008, 62, 2311–2314. [Google Scholar] [CrossRef]
- Garcia-Calvo, E.; Machuca, A.; Nerín, C.; Rosales-Conrado, N.; Anunciação, D.S.; Luque-Garcia, J.L. Integration of untargeted and targeted mass spectrometry-based metabolomics provides novel insights into the potential toxicity associated to surfynol. Food Chem. Toxicol. 2020, 146, 111849. [Google Scholar] [CrossRef] [PubMed]
- Sapcariu, S.C.; Kanashova, T.; Weindl, D.; Ghelfi, J.; Dittmar, G.; Hiller, K. Simultaneous extraction of proteins and metabolites from cells in culture. MethodsX 2014, 1, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Papadimitropoulos, M.P.; Vasilopoulou, C.G.; Maga-Nteve, C.; Klapa, M.I. Untargeted GC-MS Metabolomics. Methods Mol. Biol. 2018, 1738, 133–147. [Google Scholar] [CrossRef] [PubMed]
- Steuer, R.; Morgenthal, K.; Weckwerth, W.; Selbig, J. A gentle guide to the analysis of metabolomic data. Methods Mol. Biol. 2007, 358, 105–126. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Zhang, B.; Cheng, W.H.; Wang, Q. Preparation, characterization and evaluation of selenite-loaded chitosan/TPP nanoparticles with or without zein coating. Carbohydr. Polymers. 2010, 82, 942–951. [Google Scholar] [CrossRef]
- Zhang, S.; Luo, Y.; Zeng, H.; Wang, Q.; Tian, F.; Song, J.; Cheng, W.H. Encapsulation of selenium in chitosan nanoparticles improves selenium availability and protects cells from selenium-induced DNA damage response. J. Nutr. Biochem. 2011, 22, 1137–1142. [Google Scholar] [CrossRef]
- Rigoulet, M.; Bouchez, C.L.; Paumard, P.; Ransac, S.; Cuvellier, S.; Duvezin-Caubet, S.; Mazat, J.P.; Devin, A. Cell energy metabolism: An update. Biochim. Biophys. Acta Bioenerg. 2020, 1861, 148276. [Google Scholar] [CrossRef]
- Bonora, M.; Patergnani, S.; Rimessi, A.; De Marchi, E.; Suski, J.M.; Bononi, A.; Giorgi, C.; Marchi, S.; Missiroli, S.; Poletti, F.; et al. ATP synthesis and storage. Purinergic Signal. 2012, 8, 343–357. [Google Scholar] [CrossRef] [Green Version]
- Galber, C.; Acosta, M.J.; Minervini, G.; Giorgio, V. The role of mitochondrial ATP synthase in cancer. Biol. Chem. 2020, 401, 1199–1214. [Google Scholar] [CrossRef]
- Lunt, S.Y.; Vander Heiden, M.G. Aerobic glycolysis: Meeting the metabolic requirements of cell proliferation. Annu. Rev. Cell Dev. Biol. 2011, 27, 441–464. [Google Scholar] [CrossRef] [Green Version]
- Liberti, M.V.; Locasale, J.W. The Warburg Effect: How Does it Benefit Cancer Cells? Trends Biochem. Sci. 2016, 41, 211–218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castilla, C.; Flores, M.L.; Conde, J.M.; Medina, R.; Torrubia, F.J.; Japón, M.A.; Sáez, C. Downregulation of protein tyrosine phosphatase PTPL1 alters cell cycle and upregulates invasion-related genes in prostate cancer cells. Clin. Exp. Metastasis 2012, 29, 349–358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, S.; Jiao, W.; Xu, Y.; Hou, L.; Li, H.; Shao, J.; Zhang, X.; Wang, R.; Kong, D. Palmitic acid inhibits prostate cancer cell proliferation and metastasis by suppressing the PI3K/Akt pathway. Life Sci. 2021, 286, 120046. [Google Scholar] [CrossRef]
- DeBerardinis, R.J.; Chandel, N.S. Fundamentals of cancer metabolism. Sci. Adv. 2016, 2, e1600200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DeBerardinis, R.J. Serine metabolism: Some tumors take the road less traveled. Cell Metab. 2011, 14, 285–286. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Hancock, C.N.; Fischer, J.W.; Harman, M.; Phang, J.M. Proline biosynthesis augments tumor cell growth and aerobic glycolysis: Involvement of pyridine nucleotides. Sci. Rep. 2015, 5, 17206. [Google Scholar] [CrossRef] [Green Version]
- Geeraerts, S.L.; Heylen, E.; De Keersmaecker, K.; Kampen, K.R. The ins and outs of serine and glycine metabolism in cancer. Nat. Metab. 2021, 3, 131–141. [Google Scholar] [CrossRef] [PubMed]
- Tibbetts, A.S.; Appling, D.R. Compartmentalization of Mammalian folate-mediated one-carbon metabolism. Annu. Rev. Nutr. 2010, 30, 57–81. [Google Scholar] [CrossRef]
- Pasternack, L.B.; Laude, D.A., Jr.; Appling, D.R. 13C NMR detection of folate-mediated serine and glycine synthesis in vivo in Saccharomyces cerevisiae. Biochemistry 1992, 31, 8713–8719. [Google Scholar] [CrossRef]
- Maddocks, O.D.; Labuschagne, C.F.; Adams, P.D.; Vousden, K.H. Serine Metabolism Supports the Methionine Cycle and DNA/RNA Methylation through De Novo ATP Synthesis in Cancer Cells. Mol. Cell 2016, 61, 210–221. [Google Scholar] [CrossRef] [Green Version]
- Yang, M.; Vousden, K.H. Serine and one-carbon metabolism in cancer. Nat. Rev. Cancer 2016, 16, 650–662. [Google Scholar] [CrossRef] [PubMed]
- Kampen, K.R.; Fancello, L.; Girardi, T.; Rinaldi, G.; Planque, M.; Sulima, S.O.; Loayza-Puch, F.; Verbelen, B.; Vereecke, S.; Verbeeck, J.; et al. Translatome analysis reveals altered serine and glycine metabolism in T-cell acute lymphoblastic leukemia cells. Nat. Commun. 2019, 10, 2542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwartz, N.U.; Linzer, R.W.; Truman, J.P.; Gurevich, M.; Hannun, Y.A.; Senkal, C.E.; Obeid, L.M. Decreased ceramide underlies mitochondrial dysfunction in Charcot-Marie-Tooth 2F. FASEB J. 2018, 32, 1716–1728. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muthusamy, T.; Cordes, T.; Handzlik, M.K.; You, L.; Lim, E.W.; Gengatharan, J.; Pinto, A.; Badur, M.G.; Kolar, M.J.; Wallace, M.; et al. Serine restriction alters sphingolipid diversity to constrain tumour growth. Nature 2020, 586, 790–795. [Google Scholar] [CrossRef] [PubMed]
- Jung, K.; Reszka, R.; Kamlage, B.; Bethan, B.; Stephan, C.; Lein, M.; Kristiansen, G. Tissue metabolite profiling identifies differentiating and prognostic biomarkers for prostate carcinoma. Int. J. Cancer 2013, 133, 2914–2924. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Li, Z.; Zhang, Q.; Wei, S.; Li, B.; Zhang, X.; Zhang, L.; Li, Q.; Xu, H.; Xu, Z. Silencing of AQP3 induces apoptosis of gastric cancer cells via downregulation of glycerol intake and downstream inhibition of lipogenesis and autophagy. Onco Targets Ther. 2017, 10, 2791–2804. [Google Scholar] [CrossRef] [Green Version]
- Boison, D.; Yegutkin, G.G. Adenosine Metabolism: Emerging Concepts for Cancer Therapy. Cancer Cell. 2019, 36, 582–596. [Google Scholar] [CrossRef]
- Eniafe, J.; Jiang, S. The functional roles of TCA cycle metabolites in cancer. Oncogene 2021, 40, 3351–3363. [Google Scholar] [CrossRef]
- Zhang, B.; Tornmalm, J.; Widengren, J.; Vakifahmetoglu-Norberg, H.; Norberg, E. Characterization of the Role of the Malate Dehydrogenases to Lung Tumor Cell Survival. J. Cancer 2017, 8, 2088–2096. [Google Scholar] [CrossRef] [Green Version]
- Lim, J.K.M.; Leprivier, G. The impact of oncogenic RAS on redox balance and implications for cancer development. Cell Death Dis. 2019, 10, 955. [Google Scholar] [CrossRef] [Green Version]
- Ren, J.G.; Seth, P.; Everett, P.; Clish, C.B.; Sukhatme, V.P. Induction of erythroid differentiation in human erythroleukemia cells by depletion of malic enzyme 2. PLoS ONE 2010, 5, e12520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ren, J.G.; Seth, P.; Clish, C.B.; Lorkiewicz, P.K.; Higashi, R.M.; Lane, A.N.; Fan, T.W.; Sukhatme, V.P. Knockdown of malic enzyme 2 suppresses lung tumor growth, induces differentiation and impacts PI3K/AKT signaling. Sci. Rep. 2014, 24, 5414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peterson, C.T.; Rodionov, D.A.; Osterman, A.L.; Peterson, S.N. B Vitamins and Their Role in Immune Regulation and Cancer. Nutrients 2020, 12, 3380. [Google Scholar] [CrossRef] [PubMed]
- Pavlova, N.N.; Thompson, C.B. The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 2016, 23, 27–47. [Google Scholar] [CrossRef] [Green Version]
- Fung, M.; Chan, G.C. Drug-induced amino acid deprivation as strategy for cancer therapy. Hematol. Oncol. 2017, 10, 144. [Google Scholar] [CrossRef]
- Anderson, N.M.; Mucka, P.; Kern, J.G.; Feng, H. The emerging role and targetability of the TCA cycle in cancer metabolism. Protein Cell 2018, 9, 216–237. [Google Scholar] [CrossRef]
- Kamphorst, J.J.; Nofal, M.; Commisso, C.; Hackett, S.R.; Lu, W.; Grabocka, E.; Vander Heiden, M.G.; Miller, G.; Drebin, J.A.; Bar-Sagi, D.; et al. Human pancreatic cancer tumors are nutrient poor and tumor cells actively scavenge extracellular protein. Cancer Res. 2015, 75, 544–553. [Google Scholar] [CrossRef] [Green Version]
- Bonifácio, V.; Pereira, S.A.; Serpa, J.; Vicente, J.B. Cysteine metabolic circuitries: Druggable targets in cancer. Br. J. Cancer 2021, 124, 862–879. [Google Scholar] [CrossRef]
- Nakagawa, T.; Johnson, R.J.; Andres-Hernando, A.; Roncal-Jimenez, C.; Sanchez-Lozada, L.G.; Tolan, D.R.; Lanaspa, M.A. Fructose Production and Metabolism in the Kidney. JASN 2020, 31, 898–906. [Google Scholar] [CrossRef]
- Nakagawa, T.; Lanaspa, M.A.; Millan, I.S.; Fini, M.; Rivard, C.J.; Sanchez-Lozada, L.G.; Andres-Hernando, A.; Tolan, D.R.; Johnson, R.J. Fructose contributes to the Warburg effect for cancer growth. Cancer Metab. 2020, 8, 16. [Google Scholar] [CrossRef]
- Park, T.J.; Reznick, J.; Peterson, B.L.; Blass, G.; Omerbašić, D.; Bennett, N.C.; Kuich, P.; Zasada, C.; Browe, B.M.; Hamann, W.; et al. Fructose-driven glycolysis supports anoxia resistance in the naked mole-rat. Science 2017, 356, 307–311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Estevez, H.; Garcia-Calvo, E.; Rivera-Torres, J.; Vallet-Regí, M.; González, B.; Luque-Garcia, J.L. Transcriptome Analysis Identifies Novel Mechanisms Associated with the Antitumor Effect of Chitosan-Stabilized Selenium Nanoparticles. Pharmaceutics 2021, 13, 356. [Google Scholar] [CrossRef] [PubMed]






| Metabolite | Retention Time (min) | NIST Rmatch | RM * |
|---|---|---|---|
| Glycine | 9.56 | 844 | 0.25 |
| Butanoic acid | 9.64 | 783 | 1.84 |
| Benzoic acid | 10.14 | 718 | 1.50 |
| L-Valine | 11.55 | 848 | 0.18 |
| Serine | 12.47 | 798 | 0.61 |
| Urea | 12.63 | 825 | 1.35 |
| Glycerol | 12.91 | 896 | 3.18 |
| L-Proline | 13.33 | 729 | 0.47 |
| Pyrimidine | 14.17 | 816 | 1.50 |
| 2-butenoic acid | 14.55 | 869 | 0.21 |
| Aminomalonic acid | 17.00 | 850 | 1.81 |
| Malic acid | 17.48 | 828 | 0.15 |
| L-Cisteine | 18.71 | 905 | 3.69 |
| Glutamine | 20.06 | 844 | 0.25 |
| L-Isoleucine | 20.74 | 747 | 0.16 |
| Alpha-aminoadipic acid | 21.79 | 732 | 0.01 |
| Hydracrylic acid | 23.39 | 863 | 1.97 |
| 2-desoxigalactopiranose | 23.75 | 777 | 0.74 |
| Adenine | 24.25 | 824 | 1.20 |
| D-Fructose | 24.47 | 881 | 2.16 |
| D-Glucose | 24.89 | 817 | 1.64 |
| Pantothenic acid | 26.38 | 908 | 0.11 |
| Inositol | 26.95 | 854 | 0.22 |
| N-acetylglucosamine | 27.63 | 714 | 1.24 |
| PET-cGMP | 33.60 | 963 | 3.22 |
| 2-(1 H)-pyrimidinone | 38.15 | 805 | 0.66 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Estevez, H.; Garcia-Calvo, E.; Mena, M.L.; Alvarez-Fernandez Garcia, R.; Luque-Garcia, J.L. Unraveling the Mechanisms of Ch-SeNP Cytotoxicity against Cancer Cells: Insights from Targeted and Untargeted Metabolomics. Nanomaterials 2023, 13, 2204. https://0-doi-org.brum.beds.ac.uk/10.3390/nano13152204
Estevez H, Garcia-Calvo E, Mena ML, Alvarez-Fernandez Garcia R, Luque-Garcia JL. Unraveling the Mechanisms of Ch-SeNP Cytotoxicity against Cancer Cells: Insights from Targeted and Untargeted Metabolomics. Nanomaterials. 2023; 13(15):2204. https://0-doi-org.brum.beds.ac.uk/10.3390/nano13152204
Chicago/Turabian StyleEstevez, Hector, Estefania Garcia-Calvo, Maria L. Mena, Roberto Alvarez-Fernandez Garcia, and Jose L. Luque-Garcia. 2023. "Unraveling the Mechanisms of Ch-SeNP Cytotoxicity against Cancer Cells: Insights from Targeted and Untargeted Metabolomics" Nanomaterials 13, no. 15: 2204. https://0-doi-org.brum.beds.ac.uk/10.3390/nano13152204