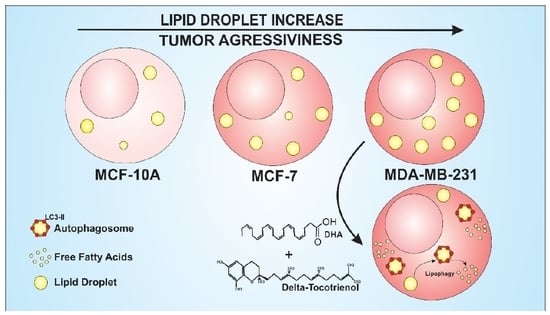
Omega 3-DHA and Delta-Tocotrienol Modulate Lipid Droplet Biogenesis and Lipophagy in Breast Cancer Cells: the Impact in Cancer Aggressiveness
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cells and Treatment
2.2. Assay of Aggressiveness—Cell Transfection with Short Interfering (si)RNA—ADRP
2.3. Western Blot
2.4. Assay of Cytotoxicity
2.5. Production of Reactive Oxygen Species (ROS)
2.6. Lipid Droplet Biogenesis Analysis by Flow Cytometry
2.7. Lipid Droplet Biogenesis Analysis by Confocal Microscopy
2.8. Lipophagy Analysis
2.9. Clonogenic Assay
2.10. Cell Migration Assay—Wound Healing Assay
2.11. Statistical Analysis
3. Results
3.1. MDA-MB-231 Cell Line Aggressiveness
3.2. Determination of DHA, Delta-T3 and Co-Treatment Cytotoxicity
3.3. Reactive Oxygen Species (ROS) Production
3.4. Lipid Droplet Biogenesis
3.5. Lipophagy Assay
3.6. Clonogenic and Cell Migration
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
References
- Baenke, F.; Peck, B.; Miess, H.; Schulze, A. Hooked on fat: The role of lipid synthesis in cancer metabolism and tumour development. Dis. Model. Mech. 2013, 6, 1353–1363. [Google Scholar] [CrossRef]
- Kuo, C.Y.; Ann, D.K. When fats commit crimes: Fatty acid metabolism, cancer stemness and therapeutic resistance. Cancer Commun. 2018, 38, 47. [Google Scholar] [CrossRef] [PubMed]
- Olzmann, J.A. Dynamics and functions of lipid droplets. Nat. Rev. Mol. Cell Biol. 2019, 20, 137–155. [Google Scholar] [CrossRef] [PubMed]
- Pol, A.; Gross, S.P.; Parton, R.G. Review: Biogenesis of the multifunctional lipid droplet: Lipids, proteins, and sites. J. Cell Biol. 2014, 204, 635–646. [Google Scholar] [CrossRef] [PubMed]
- World, S. Lipid Droplets in Cancer: Guardians of Fat in a Stressful World. Molecules 2018, 23, 1941. [Google Scholar] [Green Version]
- Abramczyk, H.; Surmacki, J.; Kopeć, M.; Olejnik, A.K.; Lubecka-Pietruszewska, K.; Fabianowska-Majewska, K. The role of lipid droplets and adipocytes in cancer. Raman imaging of cell cultures: MCF10A, MCF7, and MDA-MB-231 compared to adipocytes in cancerous human breast tissue. Analyst 2015, 140, 2224–2235. [Google Scholar] [CrossRef] [PubMed]
- Jarc, E.; Kump, A.; Malavašič, P.; Eichmann, T.O.; Zimmermann, R.; Petan, T. Lipid droplets induced by secreted phospholipase A2 and unsaturated fatty acids protect breast cancer cells from nutrient and lipotoxic stress. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2018, 1863, 247–265. [Google Scholar] [CrossRef] [PubMed]
- Accioly, M.T.; Pacheco, P.; Maya-Monteiro, C.M.; Carrossini, N.; Robbs, B.K.; Oliveira, S.S.; Kaufmann, C.; Morgado-Diaz, J.A.; Bozza, P.T.; Viola, J.P.B. Lipid Bodies Are Reservoirs of Cyclooxygenase-2 and Sites of Prostaglandin-E2 Synthesis in Colon Cancer Cells. Cancer Res. 2008, 68, 1732–1740. [Google Scholar] [CrossRef]
- Cruz, A.L.S.; Carrossini, N.; Teixeira, L.K.; Ribeiro-Pinto, L.F.; Bozza, P.T.; Viola, J.P.B. Cell Cycle Progression Regulates Biogenesis and Cellular Localization of Lipid Droplets. Mol. Cell. Biol. 2019, 39, MCB-00374. [Google Scholar] [CrossRef]
- Mitra, R.; Le, T.T.; Gorjala, P.; Goodman, O.B. Positive regulation of prostate cancer cell growth by lipid droplet forming and processing enzymes DGAT1 and ABHD5. BMC Cancer 2017, 17, 631. [Google Scholar] [CrossRef]
- Maan, M.; Peters, J.M.; Dutta, M.; Patterson, A.D. Biochemical and Biophysical Research Communications Lipid metabolism and lipophagy in cancer. Biochem. Biophys. Res. Commun. 2018, 504, 582–589. [Google Scholar] [CrossRef]
- Lettiero, B.; Inasu, M.; Kimbung, S.; Borgquist, S. Insensitivity to atorvastatin is associated with increased accumulation of intracellular lipid droplets and fatty acid metabolism in breast cancer cells. Sci. Rep. 2018, 8, 5462. [Google Scholar] [CrossRef]
- Giovannelli, P.; Di Donato, M.; Galasso, G.; Di Zazzo, E.; Bilancio, A.; Migliaccio, A. The Androgen Receptor in Breast Cancer. Front. Endocrinol. 2018, 9, 492. [Google Scholar] [CrossRef] [Green Version]
- Dai, X.; Cheng, H.; Bai, Z.; Li, J. Breast Cancer Cell Line Classification and Its Relevance with Breast Tumor Subtyping. J. Cancer 2017, 8, 3131–3141. [Google Scholar] [CrossRef] [Green Version]
- Ghoncheh, M.; Pournamdar, Z.; Salehiniya, H. Incidence and Mortality and Epidemiology of Breast Cancer in the World. Asian Pac. J. Cancer Prev. 2016, 17, 43–46. [Google Scholar] [CrossRef] [Green Version]
- Chavez, K.J.; Garimella, S.V.; Lipkowitz, S. Triple negative breast cancer cell lines: One tool in the search for better treatment of triple negative breast cancer. Breast Dis. 2011, 32, 35–48. [Google Scholar] [CrossRef] [Green Version]
- Liu, K.; Czaja, M.J. Regulation of lipid stores and metabolism by lipophagy. Cell Death Differ. 2013, 20, 3–11. [Google Scholar] [CrossRef]
- Liu, J.; Ma, D. The Role of n-3 Polyunsaturated Fatty Acids in the Prevention and Treatment of Breast Cancer. Nutrients 2014, 6, 5184–5223. [Google Scholar] [CrossRef] [Green Version]
- Fabian, C.J.; Kimler, B.F.; Hursting, S.D. Omega-3 fatty acids for breast cancer prevention and survivorship. Breast Cancer Res. 2015, 17, 62. [Google Scholar] [CrossRef]
- Blanckaert, V.; Ulmann, L.; Mimouni, V.; Antol, J.; Brancquart, L.; Chénais, B. Docosahexaenoic acid intake decreases proliferation, increases apoptosis and decreases the invasive potential of the human breast carcinoma cell line MDA-MB-231. Int. J. Oncol. 2010, 36, 737–742. [Google Scholar] [CrossRef] [Green Version]
- Sun, H.; Hu, Y.; Gu, Z.; Owens, R.T.; Chen, Y.Q.; Edwards, I.J. Omega-3 fatty acids induce apoptosis in human breast cancer cells and mouse mammary tissue through syndecan-1 inhibition of the MEK-Erk pathway. Carcinogenesis 2011, 32, 1518–1524. [Google Scholar] [CrossRef] [Green Version]
- Merendino, N.; Costantini, L.; Manzi, L.; Molinari, R.; D’Eliseo, D.; Velotti, F. Dietary ω -3 polyunsaturated fatty acid DHA: A potential adjuvant in the treatment of cancer. Biomed Res. Int. 2013, 2013, 310186. [Google Scholar] [CrossRef]
- Lecchi, C.; Invernizzi, G.; Agazzi, A.; Modina, S.; Sartorelli, P.; Savoini, G.; Ceciliani, F. Effects of EPA and DHA on lipid droplet accumulation and mRNA abundance of PAT proteins in caprine monocytes. Res. Vet. Sci. 2013, 94, 246–251. [Google Scholar] [CrossRef]
- Tiwari, R.V.; Parajuli, P.; Sylvester, P.W. γ-Tocotrienol-induced autophagy in malignant mammary cancer cells. Exp. Biol. Med. 2014, 239, 33–44. [Google Scholar] [CrossRef]
- Loganathan, R.; Selvaduray, K.R.; Nesaretnam, K.; Radhakrishnan, A.K. Tocotrienols promote apoptosis in human breast cancer cells by inducing poly(ADP-ribose) polymerase cleavage and inhibiting nuclear factor kappa-B activity. Cell Prolif. 2013, 46, 203–213. [Google Scholar] [CrossRef]
- Ramdas, P.; Rajihuzzaman, M.; Veerasenan, S.D.; Selvaduray, K.R.; Nesaretnam, K.; Radhakrishnan, A.K. Tocotrienol-treated MCF-7 human breast cancer cells show down-regulation of API5 and up-regulation of MIG6 genes. Cancer Genom. Proteom. 2011, 8, 19–31. [Google Scholar]
- Müller, L.; Theile, K.; Böhm, V. In vitro antioxidant activity of tocopherols and tocotrienols and comparison of vitamin E concentration and lipophilic antioxidant capacity in human plasma. Mol. Nutr. Food Res. 2010, 54, 731–742. [Google Scholar] [CrossRef]
- Stone, W.L.; Krishnan, K.; Campbell, S.E.; Qui, M.; Whaley, S.G.; Yang, H. Tocopherols and the Treatment of Colon Cancer. Ann. N. Y. Acad. Sci. 2004, 1031, 223–233. [Google Scholar] [CrossRef]
- Lim, S.-W.; Loh, H.-S.; Ting, K.-N.; Bradshaw, T.D.; Zeenathul, N.A. Cytotoxicity and apoptotic activities of alpha-, gamma- and delta-tocotrienol isomers on human cancer cells. BMC Complement. Altern. Med. 2014, 14, 469. [Google Scholar] [CrossRef]
- Peh, H.Y.; Tan, W.S.D.; Liao, W.; Wong, W.S.F. Vitamin E therapy beyond cancer: Tocopherol versus tocotrienol. Pharmacol. Ther. 2016, 162, 152–169. [Google Scholar] [CrossRef]
- Xiong, A.; Yu, W.; Tiwary, R.; Sanders, B.G.; Kline, K. Distinct roles of different forms of vitamin E in DHA-induced apoptosis in triple-negative breast cancer cells. Mol. Nutr. Food Res. 2012, 56, 923–934. [Google Scholar] [CrossRef]
- Almeida, J.L.; Cole, K.D.; Plant, A.L. Standards for Cell Line Authentication and Beyond. PLoS Biol. 2016, 14, e1002476. [Google Scholar] [CrossRef]
- Falagan-Lotsch, P.; Lopes, T.S.; Ferreira, N.; Balthazar, N.; Monteiro, A.M.; Borojevic, R.; Granjeiro, J.M. Performance of PCR-based and Bioluminescent assays for mycoplasma detection. J. Microbiol. Methods 2015, 118, 31–36. [Google Scholar] [CrossRef]
- Wang, C.; Ju, H.; Shen, C.; Tong, Z. miR-429 mediates δ-tocotrienol-induced apoptosis in triple-negative breast cancer cells by targeting XIAP. Int. J. Clin. Exp. Med. 2015, 8, 15648–15656. [Google Scholar]
- Pizato, N.; Luzete, B.C.; Kiffer, L.F.M.V.; Corrêa, L.H.; De Oliveira Santos, I.; Assumpção, J.A.F.; Ito, M.K.; Magalhães, K.G. Omega-3 docosahexaenoic acid induces pyroptosis cell death in triple-negative breast cancer cells. Sci. Rep. 2018, 8, 1–12. [Google Scholar]
- Shen, G.; Ning, N.; Zhao, X.; Liu, X.; Wang, G.; Wang, T.; Zhao, R.; Yang, C.; Wang, D.; Gong, P.; et al. Adipose differentiation-related protein is not involved in hypoxia inducible factor-1-induced lipid accumulation under hypoxia. Mol. Med. Rep. 2015, 12, 8055–8061. [Google Scholar] [CrossRef] [Green Version]
- Galán, J.A.; Sánchez, E.E.; Rodríguez-Acosta, A.; Soto, J.G.; Bashir, S.; McLane, M.A.; Paquette-Straub, C.; Pérez, J.C. Inhibition of lung tumor colonization and cell migration with the disintegrin crotatroxin 2 isolated from the venom of Crotalus atrox. Toxicon 2008, 51, 1186–1196. [Google Scholar] [CrossRef] [Green Version]
- Golubkov, V.; Hawes, D.; Markland, F.S. Anti-Angiogenic Activity of Contortrostatin, a Disintegrin from Agkistrodon Contortrix Contortrix Snake Venom. Angiogenesis 2003, 6, 213–224. [Google Scholar] [CrossRef]
- Yue, P.Y.K.; Leung, E.P.Y.; Mak, N.K.; Wong, R.N.S. A Simplified Method for Quantifying Cell Migration/Wound Healing in 96-Well Plates. J. Biomol. Screen. 2010, 15, 427–433. [Google Scholar] [CrossRef] [Green Version]
- Marmé, F.; Schneeweiss, A. Targeted Therapies in Triple-Negative Breast Cancer. Breast Care 2015, 10, 159–166. [Google Scholar] [CrossRef]
- Xue, M.; Ge, Y.; Yu, C.; Zheng, Z.; He, X.; Zhao, J. Apoptosis is induced by docosahexaenoic acid in breast cancer cells via death receptor and mitochondria-mediated pathways. Mol. Med. Rep. 2017, 16, 978–982. [Google Scholar] [CrossRef] [PubMed]
- Abramczyk, H.; Brozek-Pluska, B.; Surmacki, J.; Jablonska-Gajewicz, J.; Kordek, R. Raman ‘optical biopsy’ of human breast cancer. Prog. Biophys. Mol. Biol. 2012, 108, 74–81. [Google Scholar] [CrossRef] [PubMed]
- Wright, H.J.; Hou, J.; Xu, B.; Cortez, M.; Potma, E.O.; Tromberg, B.J.; Razorenova, O.V. CDCP1 drives triple-negative breast cancer metastasis through reduction of lipid-droplet abundance and stimulation of fatty acid oxidation. Proc. Natl. Acad. Sci. USA 2017, 114, E6556–E6565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sunami, Y.; Rebelo, A.; Kleeff, J. Lipid Metabolism and Lipid Droplets in Pancreatic Cancer and Stellate Cells. Cancers 2017, 10, 3. [Google Scholar] [CrossRef] [PubMed]
- Ambrosio, M.R.; Piccaluga, P.P.; Ponzoni, M.; Rocca, B.J.; Malagnino, V.; Onorati, M.; De Falco, G.; Calbi, V.; Ogwang, M.; Naresh, K.N.; et al. The Alteration of Lipid Metabolism in Burkitt Lymphoma Identifies a Novel Marker: Adipophilin. PLoS ONE 2012, 7, e44315. [Google Scholar] [CrossRef] [PubMed]
- Mitra, R.; Chao, O.; Urasaki, Y.; Goodman, O.B.; Le, T.T. Detection of Lipid-Rich Prostate Circulating Tumour Cells with Coherent Anti-Stokes Raman Scattering Microscopy. BMC Cancer 2012, 12, 540. [Google Scholar] [CrossRef]
- Nakagawa, H.; Hayata, Y.; Kawamura, S.; Yamada, T.; Fujiwara, N.; Koike, K. Lipid Metabolic Reprogramming in Hepatocellular Carcinoma. Cancers 2018, 10, 447. [Google Scholar] [CrossRef]
- Koizume, S.; Miyagi, Y. Lipid Droplets: A Key Cellular Organelle Associated with Cancer Cell Survival under Normoxia and Hypoxia. Int. J. Mol. Sci. 2016, 17, 1430. [Google Scholar] [CrossRef]
- Tirinato, L.; Pagliari, F.; Limongi, T.; Marini, M.; Falqui, A.; Seco, J.; Candeloro, P.; Liberale, C.; Di Fabrizio, E. An Overview of Lipid Droplets in Cancer and Cancer Stem Cells. Stem Cells Int. 2017, 2017, 1–17. [Google Scholar] [CrossRef]
- El-Mashtoly, S.F.; Yosef, H.K.; Petersen, D.; Mavarani, L.; Maghnouj, A.; Hahn, S.; Kötting, C.; Gerwert, K. Label-Free Raman Spectroscopic Imaging Monitors the Integral Physiologically Relevant Drug Responses in Cancer Cells. Anal. Chem. 2015, 87, 7297–7304. [Google Scholar] [CrossRef]
- Rak, S.; De Zan, T.; Stefulj, J.; Kosović, M.; Gamulin, O.; Osmak, M. FTIR spectroscopy reveals lipid droplets in drug resistant laryngeal carcinoma cells through detection of increased ester vibrational bands intensity. Analyst 2014, 139, 3407–3415. [Google Scholar] [CrossRef]
- Jarc, E.; Eichmann, T.O.; Zimmermann, R.; Petan, T. Lipidomic data on lipid droplet triglyceride remodelling associated with protection of breast cancer cells from lipotoxic stress. Data Br. 2018, 18, 234–240. [Google Scholar] [CrossRef]
- Walch, L.; Čopič, A.; Jackson, C.L. Fatty Acid Metabolism Meets Organelle Dynamics. Dev. Cell 2015, 32, 657–658. [Google Scholar] [CrossRef] [Green Version]
- Listenberger, L.L.; Han, X.; Lewis, S.E.; Cases, S.; Farese, R.V.; Ory, D.S.; Schaffer, J.E. Triglyceride accumulation protects against fatty acid-induced lipotoxicity. Proc. Natl. Acad. Sci. USA 2003, 100, 3077–3082. [Google Scholar] [CrossRef] [Green Version]
- Bozza, P.T.; Viola, J.P.B. Lipid droplets in inflammation and cancer. Prostaglandins Leukot. Essent. Fat. Acids 2010, 82, 243–250. [Google Scholar] [CrossRef]
- Currie, E.; Schulze, A.; Zechner, R.; Walther, T.C.; Farese, R.V. Cellular Fatty Acid Metabolism and Cancer. Cell Metab. 2013, 18, 153–161. [Google Scholar] [CrossRef] [Green Version]
- Xu, H.N.; Nioka, S.; Glickson, J.D.; Chance, B.; Li, L.Z. Quantitative mitochondrial redox imaging of breast cancer metastatic potential. J. Biomed. Opt. 2010, 15, 036010. [Google Scholar] [CrossRef]
- Zhang, C.; Yu, H.; Shen, Y.; Ni, X.; Shen, S.; Das, U.N. Polyunsaturated fatty acids trigger apoptosis of colon cancer cells through a mitochondrial pathway. Arch. Med. Sci. 2015, 11, 1081–1094. [Google Scholar]
- Burdeos, G.C.; Nakagawa, K.; Abe, T.; Kimura, F.; Miyazawa, T. Tocotrienol modulates crucial lipid metabolism-related genes in differentiated 3T3-L1 preadipocytes. Food Funct. 2014, 5, 2221–2227. [Google Scholar] [CrossRef]
- Cardenas, E.; Ghosh, R. Vitamin E: A dark horse at the crossroad of cancer management. Biochem. Pharmacol. 2013, 86, 845–852. [Google Scholar] [CrossRef] [Green Version]
- Aggarwal, B.B.; Sundaram, C.; Prasad, S.; Kannappan, R. Tocotrienols, the vitamin E of the 21st century: Its potential against cancer and other chronic diseases. Biochem. Pharmacol. 2010, 80, 1613–1631. [Google Scholar] [CrossRef] [Green Version]
- Wong, R.S.; Radhakrishnan, A.K. Tocotrienol research: Past into present. Nutr. Rev. 2012, 70, 483–490. [Google Scholar] [CrossRef]
- Mizushima, N.; Levine, B. Autophagy in mammalian development and differentiation. Nat. Cell Biol. 2010, 12, 823–830. [Google Scholar] [CrossRef]
- White, E. The role for autophagy in cancer. J. Clin. Invest. 2015, 125, 42–46. [Google Scholar] [CrossRef] [Green Version]
- Mathew, R.; White, E. Autophagy in tumorigenesis and energy metabolism: Friend by day, foe by night. Curr. Opin. Genet. Dev. 2011, 21, 113–119. [Google Scholar] [CrossRef]
- Rabinowitz, J.D.; White, E. Autophagy and Metabolism. Science 2010, 330, 1344–1348. [Google Scholar] [CrossRef] [Green Version]
- Singh, R.; Kaushik, S.; Wang, Y.; Xiang, Y.; Novak, I.; Komatsu, M.; Tanaka, K.; Cuervo, A.M.; Czaja, M.J. Autophagy regulates lipid metabolism. Nature 2009, 458, 1131–1135. [Google Scholar] [CrossRef] [Green Version]
- Pereira, L.; Girardi, J.P.; Bakovic, M. Forms, Crosstalks, and the Role of Phospholipid Biosynthesis in Autophagy. Int. J. Cell Biol. 2012, 2012, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Dupont, N.; Chauhan, S.; Arko-Mensah, J.; Castillo, E.F.; Masedunskas, A.; Weigert, R.; Robenek, H.; Proikas-Cezanne, T.; Deretic, V. Neutral lipid stores and lipase PNPLA5 contribute to autophagosome biogenesis. Curr. Biol. 2014, 24, 609–620. [Google Scholar] [CrossRef]
- Krahmer, N.; Farese, R.V.; Walther, T.C.; Walther, T.C. Balancing the fat: Lipid droplets and human disease. EMBO Mol. Med. 2013, 5, 973–983. [Google Scholar] [CrossRef]
- Cohen, B.-C.; Shamay, A.; Argov-Argaman, N. Regulation of Lipid Droplet Size in Mammary Epithelial Cells by Remodeling of Membrane Lipid Composition—A Potential Mechanism. PLoS ONE 2015, 10, e0121645. [Google Scholar] [CrossRef] [PubMed]
- Chitraju, C.; Trötzmüller, M.; Hartler, J.; Wolinski, H.; Thallinger, G.G.; Lass, A.; Zechner, R.; Zimmermann, R.; Köfeler, H.C.; Spener, F. Lipidomic analysis of lipid droplets from murine hepatocytes reveals distinct signatures for nutritional stress. J. Lipid Res. 2012, 53, 2141–2152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- D’Andrea, S. Lipid droplet mobilization: The different ways to loosen the purse strings. Biochimie 2016, 120, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, M.; Waguri, S.; Ueno, T.; Iwata, J.; Murata, S.; Tanida, I.; Ezaki, J.; Mizushima, N.; Ohsumi, Y.; Uchiyama, Y.; et al. Impairment of starvation-induced and constitutive autophagy in Atg7 -deficient mice. J. Cell Biol. 2005, 169, 425–434. [Google Scholar] [CrossRef] [PubMed]
- Mei, S.; Ni, H.-M.; Manley, S.; Bockus, A.; Kassel, K.M.; Luyendyk, J.P.; Copple, B.L.; Ding, W.-X. Differential Roles of Unsaturated and Saturated Fatty Acids on Autophagy and Apoptosis in Hepatocytes. J. Pharmacol. Exp. Ther. 2011, 339, 487–498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niso-Santano, M.; Malik, S.A.; Pietrocola, F.; Bravo-San Pedro, J.M.; Marino, G.; Cianfanelli, V.; Ben-Younes, A.; Troncoso, R.; Markaki, M.; Sica, V.; et al. Unsaturated fatty acids induce non-canonical autophagy. EMBO J. 2015, 34, 1025–1041. [Google Scholar] [CrossRef]
- Ahsan, H.; Ahad, A.; Iqbal, J.; Siddiqui, W.A. Pharmacological potential of tocotrienols: A review. Nutr. Metab. 2014, 11, 52. [Google Scholar] [CrossRef]
- Kline, K.; Yu, W.; Sanders, B.G. Vitamin E and Breast Cancer. J. Nutr. 2004, 134, 3458S–3462S. [Google Scholar] [CrossRef] [Green Version]
- Cheng, G.; Zielonka, J.; McAllister, D.M.; Mackinnon, A.C.; Joseph, J.; Dwinell, M.B.; Kalyanaraman, B. Mitochondria-targeted vitamin E analogs inhibit breast cancer cell energy metabolism and promote cell death. BMC Cancer 2013, 13, 285. [Google Scholar] [CrossRef]
- Siveen, K.S.; Ahn, K.S.; Ong, T.H.; Shanmugam, M.K.; Li, F.; Yap, W.N.; Kumar, A.P.; Fong, C.W.; Tergaonkar, V.; Hui, K.M.; et al. γ-tocotrienol inhibits angiogenesis-dependent growth of human hepatocellular carcinoma through abrogation of AKT/mTOR pathway in an orthotopic mouse model. Oncotarget 2014, 5, 1897–1911. [Google Scholar] [CrossRef]
- Rajendran, P.; Li, F.; Manu, K.A.; Shanmugam, M.K.; Loo, S.Y.; Kumar, A.P.; Sethi, G. γ-Tocotrienol is a novel inhibitor of constitutive and inducible STAT3 signalling pathway in human hepatocellular carcinoma: Potential role as an antiproliferative, pro-apoptotic and chemosensitizing agent. Br. J. Pharmacol. 2011, 163, 283–298. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Yin, X.; Jiang, Q. Natural Forms of Vitamin E and 13′-Carboxychromanol, a Long-Chain Vitamin E Metabolite, Inhibit Leukotriene Generation from Stimulated Neutrophils by Blocking Calcium Influx and Suppressing 5-Lipoxygenase Activity, Respectively. J. Immunol. 2011, 186, 1173–1179. [Google Scholar] [CrossRef] [PubMed]
- Uto-Kondo, H.; Ohmori, R.; Kiyose, C.; Kishimoto, Y.; Saito, H.; Igarashi, O.; Kondo, K. Tocotrienol Suppresses Adipocyte Differentiation and Akt Phosphorylation in 3T3-L1 Preadipocytes. J. Nutr. 2009, 139, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Burdeos, G.C.; Nakagawa, K.; Kimura, F.; Miyazawa, T. Tocotrienol Attenuates Triglyceride Accumulation in HepG2 Cells and F344 Rats. Lipids 2012, 47, 471–481. [Google Scholar] [CrossRef] [PubMed]
- Kaini, R.R.; Sillerud, L.O.; Zhaorigetu, S.; Hu, C.-A.A. Autophagy regulates lipolysis and cell survival through lipid droplet degradation in androgen-sensitive prostate cancer cells. Prostate 2012, 72, 1412–1422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tran, A.T.; Ramalinga, M.; Kedir, H.; Clarke, R.; Kumar, D. Autophagy inhibitor 3-methyladenine potentiates apoptosis induced by dietary tocotrienols in breast cancer cells. Eur. J. Nutr. 2015, 54, 265–272. [Google Scholar] [CrossRef]
- Sun, S.-N.; Jia, W.-D.; Chen, H.; Ma, J.-L.; Ge, Y.-S.; Yu, J.-H.; Li, J.-S. Docosahexaenoic acid (DHA) induces apoptosis in human hepatocellular carcinoma cells. Int. J. Clin. Exp. Pathol. 2013, 6, 281–289. [Google Scholar] [PubMed]
- Wu, M.; Harvey, K.A.; Ruzmetov, N.; Welch, Z.R.; Sech, L.; Jackson, K.; Stillwell, W.; Zaloga, G.P.; Siddiqui, R.A. Omega-3 polyunsaturated fatty acids attenuate breast cancer growth through activation of a neutral sphingomyelinase-mediated pathway. Int. J. Cancer 2005, 117, 340–348. [Google Scholar] [CrossRef]
- Kang, K.S.; Wang, P.; Yamabe, N.; Fukui, M.; Jay, T.; Zhu, B.T. Docosahexaenoic acid induces apoptosis in MCF-7 cells in vitro and in vivo via reactive oxygen species formation and caspase 8 activation. PLoS ONE 2010, 5, e10296. [Google Scholar] [CrossRef]
- Blanckaert, V.; Kerviel, V.; Lépinay, A.; Joubert-Durigneux, V.; Hondermarck, H.; Chénais, B. Docosahexaenoic acid inhibits the invasion of MDA-MB-231 breast cancer cells through upregulation of cytokeratin-1. Int. J. Oncol. 2015, 46, 2649–2655. [Google Scholar] [CrossRef]
- Shin, S.; Jing, K.; Jeong, S.; Kim, N.; Song, K.; Heo, J.; Park, J.; Seo, K.; Han, J.; Park, J.; et al. The Omega-3 Polyunsaturated Fatty Acid DHA Induces Simultaneous Apoptosis and Autophagy via Mitochondrial ROS-Mediated Akt-mTOR Signaling in Prostate Cancer Cells Expressing Mutant p53. Biomed Res. Int. 2013, 2013, 568671. [Google Scholar] [CrossRef] [PubMed]
- Guièze, R.; Gyan, E.; Tournilhac, O.; Halty, C.; Veyrat-Masson, R.; Akil, S.; Berger, M.; Hérault, O.; Callanan, M.; Bay, J.-O. Docosahexaenoic Acid Induces Apoptosis in Primary Chronic Lymphocytic Leukemia Cells. Hematol. Rep. 2015, 7, 6043. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kolar, S.; Barhoumi, R.; Jones, C.K.; Wesley, J.; Lupton, J.R.; Fan, Y.-Y.; Chapkin, R.S. Interactive effects of fatty acid and butyrate-induced mitochondrial Ca2+ loading and apoptosis in colonocytes. Cancer 2011, 117, 5294–5303. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, K.; Monsen, V.T.; Hakvåg Pettersen, C.H.; Overland, H.B.; Pettersen, G.; Samdal, H.; Tesfahun, A.N.; Lundemo, A.G.; Bjørkøy, G.; Schønberg, S.A. DHA-induced stress response in human colon cancer cells—Focus on oxidative stress and autophagy. Free Radic. Biol. Med. 2016, 90, 158–172. [Google Scholar] [CrossRef] [PubMed]
- Strouch, M.J.; Ding, Y.; Salabat, M.R.; Melstrom, L.G.; Adrian, K.; Quinn, C.; Pelham, C.; Rao, S.; Adrian, T.E.; Bentrem, D.J.; et al. A High Omega-3 Fatty Acid Diet Mitigates Murine Pancreatic Precancer Development. J. Surg. Res. 2011, 165, 75–81. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Antalis, C.J.; Uchida, A.; Buhman, K.K.; Siddiqui, R.A. Migration of MDA-MB-231 breast cancer cells depends on the availability of exogenous lipids and cholesterol esterification. Clin. Exp. Metastasis 2011, 28, 733–741. [Google Scholar] [CrossRef] [PubMed]






© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pizato, N.; Kiffer, L.F.M.V.; Luzete, B.C.; Assumpção, J.A.F.; Correa, L.H.; Melo, H.A.B.d.; Sant’Ana, L.P.d.; Ito, M.K.; Magalhães, K.G. Omega 3-DHA and Delta-Tocotrienol Modulate Lipid Droplet Biogenesis and Lipophagy in Breast Cancer Cells: the Impact in Cancer Aggressiveness. Nutrients 2019, 11, 1199. https://0-doi-org.brum.beds.ac.uk/10.3390/nu11061199
Pizato N, Kiffer LFMV, Luzete BC, Assumpção JAF, Correa LH, Melo HABd, Sant’Ana LPd, Ito MK, Magalhães KG. Omega 3-DHA and Delta-Tocotrienol Modulate Lipid Droplet Biogenesis and Lipophagy in Breast Cancer Cells: the Impact in Cancer Aggressiveness. Nutrients. 2019; 11(6):1199. https://0-doi-org.brum.beds.ac.uk/10.3390/nu11061199
Chicago/Turabian StylePizato, Nathalia, Larissa Fernanda Melo Vasconcelos Kiffer, Beatriz Christina Luzete, José Antonio Fagundes Assumpção, Luis Henrique Correa, Heloisa Antoniella Braz de Melo, Lívia Pimentel de Sant’Ana, Marina Kiyomi Ito, and Kelly Grace Magalhães. 2019. "Omega 3-DHA and Delta-Tocotrienol Modulate Lipid Droplet Biogenesis and Lipophagy in Breast Cancer Cells: the Impact in Cancer Aggressiveness" Nutrients 11, no. 6: 1199. https://0-doi-org.brum.beds.ac.uk/10.3390/nu11061199