Microbiota and Diabetes Mellitus: Role of Lipid Mediators
Abstract
:1. Introduction
2. Chronic Inflammation in Diabetes: Who Are the Precursors?
3. Microbiota
4. Microbiota and Diabetes: Immunomodulatory Role of Bacterial Lipid Mediators
4.1. Release of Lipopolysaccharides
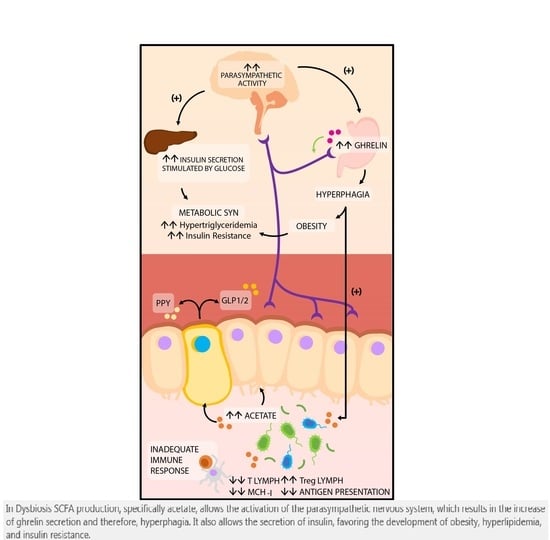
4.2. Production of Short Chain Fatty Acids
5. Microbiota and Diabetes: Therapeutic Aspects
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| DM | Diabetes Mellitus |
| LPS | Lipopolysaccharides |
| SCFA | Short Chain Fatty Acids |
| IR | Insulin Resistance |
| LBP | Lipopolysaccharide Binding Proteins |
| TLR-4 | Toll-Like Receptors 4 |
| FAK | Focal Adhesion Kinase |
| MyD88 | Myeloid Differentiation Gene 88 |
| IRAK4 | Interleukin 1 Receptor Associated Kinase 4 |
| Th17 | T Helper 17 Lymphocytes |
| TNF-α | Tumor Necrosis Factor Alfa |
| NF-ΚB | Nuclear Factor Kappa B |
| PDX-1 | Pancreatic and Duodenal Homeobox 1 |
| IRS | Insulin Receptor Substrate |
| IRβ | Insulin Receptor Beta |
| DCs | Dendritic Cells |
| IgA | Immunoglobulin A |
| GLP | Glucagon-Like Peptide |
| GP43 | Protein G43 Coupled Receptors |
| MCH-I | Type I Major Histocompatibility |
| Treg | Regulatory T Cells |
| TMAO | N-Oxide of Trimethylamine |
| AMPK | Activated Protein Kinase |
| BSH | Bile Salt Hydrolase Enzyme |
| GUDCA | Glycoursodeoxycholic Acid |
| FXR | Farnesoid X Receptor |
| CPR | C-reactive protein |
| IL | Interleukin |
| TGF-β | Transforming Growth Factor β |
| FFA | Free Fatty Acids Receptors |
| GPCR | G-protein coupled receptors |
| ILC3 | Innate lymphoid cells of the group 3 |
| HDAC | Histone Deacetylases |
| NOS2 | Nitric Oxide Synthase 2 |
| PPY | YY Peptide |
| NPY | Neuropeptide Y |
| POMC | Proopiomelanocortin |
References
- International Diabetes Federation. IDF Diabetes Atlas. 2015. Available online: https://www.idf.org/elibrary/epidemiology-research/diabetes-atlas/13-diabetes-atlas-seventh-edition.html (accessed on 20 March 2020).
- Ministery of Health. Bolivarian Republic of Venezuela. Anuary of Morbility. 2011. Available online: https://www.ovsalud.org/descargas/publicaciones/documentos-oficiales/Anuario-Morbilidad-2011.pdf (accessed on 20 March 2020).
- World Health Organization. Global Report on Diabetes; World Health Organization: Geneva, Switzerland, 2016; Available online: http://apps.who.int/iris/bitstream/10665/204871/1/9789241565257_eng.pdf (accessed on 20 March 2020).
- De La Cruz Vargas, J.A.; Dos Santos, F.; Dyzinger, W.; Herzog, S. Medicina Del Estilo de Vida: Trabajando Juntos Para Revertir La Epidemia de Las Enfermedades Crónicas En Latinoamérica. Cienc. Innov. Salud 2017, 4. [Google Scholar] [CrossRef] [Green Version]
- Baratieri, T.; Dal Santo Ottoni, J.; Luciana Botti, M.; Serpa Maicel, R.D.C.; Gramazio Soares, L. Risco Cardiovascular Em Usuários de Programa de Atenção a Hipertensos e Diabéticos Em Um Município Do Paraná-Brasil. Cienc. Innov. Salud 2014, 2. [Google Scholar] [CrossRef]
- Morales, J.; Carcausto, W.; Varillas, Y.; Pérez, J.; Salsavilca, E.; Castro, I.; Rivera, M.; Quispe, M. Actividad Física En Pacientes Con Diabetes Mellitus Del Primer Nivel de Atención de Lima Norte. Rev. Latinoam. Hipertens. 2018, 13, 49–54. [Google Scholar]
- De Fronzo, R.A. From the Triumvirate to the Ominous Octet: A New Paradigm for the Treatment of Type 2 Diabetes Mellitus. Diabetes 2009, 58, 773–795. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwartz, S.S.; Epstein, S.; Corkey, B.E.; Grant, S.F.A.; Gavin, J.R.; Aguilar, R.B. The Time Is Right for a New Classification System for Diabetes: Rationale and Implications of the β-Cell–Centric Classification Schema. Diabetes Care 2016, 39, 179–186. [Google Scholar] [CrossRef] [Green Version]
- Vatanen, T.; Kostic, A.D.; D’Hennezel, E.; Siljander, H.; Franzosa, E.A.; Yassour, M.; Kolde, R.; Vlamakis, H.; Arthur, T.D.; Hämäläinen, A.-M.; et al. Variation in Microbiome LPS Immunogenicity Contributes to Autoimmunity in Humans. Cell 2016, 165, 842–853. [Google Scholar] [CrossRef] [Green Version]
- Brown, K.; Godovannyi, A.; Ma, C.; Zhang, Y.; Ahmadi-Vand, Z.; Dai, C.; Gorzelak, M.A.; Chan, Y.; Chan, J.M.; Lochner, A.; et al. Prolonged Antibiotic Treatment Induces a Diabetogenic Intestinal Microbiome That Accelerates Diabetes in NOD Mice. ISME J. 2016, 10, 321–332. [Google Scholar] [CrossRef]
- Palacios, T.; Vitetta, L.; Coulson, S.; Madigan, C.D.; Denyer, G.S.; Caterson, I.D. The Effect of a Novel Probiotic on Metabolic Biomarkers in Adults with Prediabetes and Recently Diagnosed Type 2 Diabetes Mellitus: Study Protocol for a Randomized Controlled Trial. Trials 2017, 18. [Google Scholar] [CrossRef] [Green Version]
- Peng, J.; Narasimhan, S.; Marchesi, J.R.; Benson, A.; Wong, F.S.; Wen, L. Long Term Effect of Gut Microbiota Transfer on Diabetes Development. J. Autoimmun. 2014, 53, 85–94. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez, C.M.C.; Quiroz, E.A.N.; Lastre-Amell, G.; Oróstegui-Santander, M.A.; Peña, G.E.G.; Sucerquia, A.; Carrero, L.L.S. Dislipidemia como factor de riesgo cardiovascular: Uso de probióticos en la terapéutica nutricional. Arch. Venez. Farmacol. Ter. 2020, 39, 126–139. [Google Scholar]
- Herder, C.; Færch, K.; Carstensen-Kirberg, M.; Lowe, G.D.; Haapakoski, R.; Witte, D.R.; Brunner, E.J.; Roden, M.; Tabák, A.G.; Kivimäki, M.; et al. Biomarkers of Subclinical Inflammation and Increases in Glycaemia, Insulin Resistance and Beta-Cell Function in Non-Diabetic Individuals: The Whitehall II Study. Eur. J. Endocrinol. 2016, 175, 367–377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pouvreau, C.; Dayre, A.; Butkowski, E.G.; de Jong, B.; Jelinek, H.F. Inflammation and Oxidative Stress Markers in Diabetes and Hypertension. J. Inflamm. Res. 2018, 11, 61–68. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Odegaard, A.O.; Jacobs, D.R.; Sanchez, O.A.; Goff, D.C.; Reiner, A.P.; Gross, M.D. Oxidative Stress, Inflammation, Endothelial Dysfunction and Incidence of Type 2 Diabetes. Cardiovasc. Diabetol. 2016, 15, 51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bermudez, V.; Salazar, J.; Gonzalez, R.; Ortega, A.; Calvo, M.; Olivar, L.C.; Morillo, J.; Miquilena, E.; Chavez-Castillo, M.; Chaparro, R.; et al. Prevalence and Risk Factors Associated with Impaired Fasting Glucose in Adults from Maracaibo City, Venezuela. J. Diabetes Metab. 2019, 7, 100683. [Google Scholar] [CrossRef]
- Rojas, J.; Bermudez, V.; Palmar, J.; Martínez, M.S.; Olivar, L.C.; Nava, M.; Tomey, D.; Rojas, M.; Salazar, J.; Garicano, C.; et al. Pancreatic Beta Cell Death: Novel Potential Mechanisms in Diabetes Therapy. J. Diabetes Res. 2018, 2018, 9601801. [Google Scholar] [CrossRef] [Green Version]
- Mobini, R.; Tremaroli, V.; Ståhlman, M.; Karlsson, F.; Levin, M.; Ljungberg, M.; Sohlin, M.; Bertéus Forslund, H.; Perkins, R.; Bäckhed, F.; et al. Metabolic Effects of Lactobacillus Reuteri DSM 17938 in People with Type 2 Diabetes: A Randomized Controlled Trial. Diabetes Obes. Metab. 2017, 19, 579–589. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez Nieves, R.R.; Torres Ruiz, L.E.; Sarmiento Segarra, K.B.; Narea Illescas, D.I.; Araque Pluas, I.V.; Apolo Montero, A.M.; Ibarra Vélez, L.S.; Alvarado Chiquito, O.L. Prevalencia de Síndrome Metabólico En Trabajadores de Una Empresa de Construcción En Guayaquil, Ecuador. Rev. Latinoam. Hipertens. 2019, 14, 638–643. [Google Scholar]
- Ahmad, R.; Thomas, R.; Kochumon, S.; Sindhu, S. Increased Adipose Tissue Expression of IL-18R and Its Ligand IL-18 Associates with Inflammation and Insulin Resistance in Obesity. Immun. Inflamm. Dis. 2017, 5, 318–335. [Google Scholar] [CrossRef]
- Yan, Y.; Li, S.; Liu, Y.; Bazzano, L.; He, J.; Mi, J.; Chen, W. Temporal Relationship between Inflammation and Insulin Resistance and Their Joint Effect on Hyperglycemia: The Bogalusa Heart Study. Cardiovasc. Diabetol. 2019, 18, 109. [Google Scholar] [CrossRef] [Green Version]
- Esser, N.; Legrand-Poels, S.; Piette, J.; Scheen, A.J.; Paquot, N. Inflammation as a Link between Obesity, Metabolic Syndrome and Type 2 Diabetes. Diabetes Res. Clin. Pract. 2014, 105, 141–150. [Google Scholar] [CrossRef] [Green Version]
- Zozulinska, D.; Wierusz-Wysocka, B. Type 2 Diabetes Mellitus as Inflammatory Disease. Diabetes Res. Clin. Pract. 2006, 74, S12–S16. [Google Scholar] [CrossRef]
- Roohi, A.; Tabrizi, M.; Abbasi, F.; Ataie-Jafari, A.; Nikbin, B.; Larijani, B.; Qorbani, M.; Meysamie, A.; Asgarian-Omran, H.; Nikmanesh, B.; et al. Serum IL-17, IL-23, and TGF-β Levels in Type 1 and Type 2 Diabetic Patients and Age-Matched Healthy Controls. Biomed. Res. Int. 2014, 2014, 718946. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdel-Moneim, A.; Bakery, H.H.; Allam, G. The Potential Pathogenic Role of IL-17/Th17 Cells in Both Type 1 and Type 2 Diabetes Mellitus. Biomed. Pharmacother. 2018, 101, 287–292. [Google Scholar] [CrossRef] [PubMed]
- Von Scholten, B.J.; Reinhard, H.; Hansen, T.W.; Schalkwijk, C.G.; Stehouwer, C.; Parving, H.-H.; Jacobsen, P.K.; Rossing, P. Markers of Inflammation and Endothelial Dysfunction Are Associated with Incident Cardiovascular Disease, All-Cause Mortality, and Progression of Coronary Calcification in Type 2 Diabetic Patients with Microalbuminuria. J. Diabetes Complicat. 2016, 30, 248–255. [Google Scholar] [CrossRef]
- Sigurdardottir, S.; Zapadka, T.E.; Lindstrom, S.I.; Liu, H.; Taylor, B.E.; Lee, C.A.; Kern, T.S.; Taylor, P.R. Diabetes-Mediated IL-17A Enhances Retinal Inflammation, Oxidative Stress, and Vascular Permeability. Cell. Immunol. 2019, 341, 103921. [Google Scholar] [CrossRef]
- Román-Pintos, L.M.; Villegas-Rivera, G.; Rodríguez-Carrizalez, A.D.; Miranda-Díaz, A.G.; Cardona-Muñoz, E.G. Diabetic Polyneuropathy in Type 2 Diabetes Mellitus: Inflammation, Oxidative Stress, and Mitochondrial Function. J. Diabetes Res. 2016, 2016, 3425617. [Google Scholar] [CrossRef] [Green Version]
- Burcelin, R. Gut Microbiota and Immune Crosstalk in Metabolic Disease. Mol. Metab. 2016, 5, 771–781. [Google Scholar] [CrossRef]
- Huang, X.; Yan, D.; Xu, M.; Li, F.; Ren, M.; Zhang, J.; Wu, M. Interactive Association of Lipopolysaccharide and Free Fatty Acid with the Prevalence of Type 2 Diabetes: A Community-Based Cross-Sectional Study. J. Diabetes Investig. 2019, 10, 1438–1446. [Google Scholar] [CrossRef] [Green Version]
- Khondkaryan, L.; Margaryan, S.; Poghosyan, D.; Manukyan, G. Impaired Inflammatory Response to LPS in Type 2 Diabetes Mellitus. Int. J. Inflam. 2018, 2018, 2157434. [Google Scholar] [CrossRef] [Green Version]
- Zhao, L.; Zhang, F.; Ding, X.; Wu, G.; Lam, Y.Y.; Wang, X.; Fu, H.; Xue, X.; Lu, C.; Ma, J.; et al. Gut Bacteria Selectively Promoted by Dietary Fibers Alleviate Type 2 Diabetes. Science 2018, 359, 1151–1156. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; van Esch, B.C.A.M.; Wagenaar, G.T.M.; Garssen, J.; Folkerts, G.; Henricks, P.A.J. Pro- and Anti-Inflammatory Effects of Short Chain Fatty Acids on Immune and Endothelial Cells. Eur. J. Pharmacol. 2018, 831, 52–59. [Google Scholar] [CrossRef] [PubMed]
- Turnbaugh, P.J.; Ley, R.E.; Hamady, M.; Fraser-Liggett, C.M.; Knight, R.; Gordon, J.I. The Human Microbiome Project. Nature 2007, 449, 804–810. [Google Scholar] [CrossRef] [PubMed]
- Martí, J.M.; Martínez-Martínez, D.; Rubio, T.; Gracia, C.; Peña, M.; Latorre, A.; Moya, A.; Garay, C.P. Health and Disease Imprinted in the Time Variability of the Human Microbiome. MSystems 2017, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romero, G. Influencia de La Microbiota Intestinal En La Enfermedad Hepática Crónica. Su Rol En El Hepatocarcinoma. Gen 2016, 70, 64–69. [Google Scholar]
- Vargas-Robles, D.D.; Domínguez-Bello, M.G. Microbiota de los indígenas del Amazonas venezolano: Influencia de los estilos de vida. Gac. Med. Caracas 2020, 126, 291–303. [Google Scholar]
- Hill, C.J.; Lynch, D.B.; Murphy, K.; Ulaszewska, M.; Jeffery, I.B.; O’Shea, C.A.; Watkins, C.; Dempsey, E.; Mattivi, F.; Tuohy, K.; et al. Evolution of Gut Microbiota Composition from Birth to 24 Weeks in the INFANTMET Cohort. Microbiome 2017, 5, 4. [Google Scholar] [CrossRef] [Green Version]
- Torres, Y.; Bermúdez, V.; Garicano, C.; Vilasmil, N.; Bautista, J.; Martínez, M.S.; Rojas-Quintero, J. Desarrollo del sistema inmunológico ¿naturaleza o crianza? Arch. Venez. Farmacol. Ter. 2017, 36, 144–151. [Google Scholar]
- Faneite Antique, D.P.; Faneite Campos, J. Microbioma perinatal: Nuevos horizontes de la vida. Gac. Med. Caracas 2020, 123, 94–106. [Google Scholar]
- Duranti, S.; Lugli, G.A.; Milani, C.; James, K.; Mancabelli, L.; Turroni, F.; Alessandri, G.; Mangifesta, M.; Mancino, W.; Ossiprandi, M.C.; et al. Bifidobacterium bifidum and the infant gut microbiota: An intriguing case of microbe-host co-evolution. Environ. Microbiol. 2019, 21, 3683–3695. [Google Scholar] [CrossRef]
- Biagi, E.; Nylund, L.; Candela, M.; Ostan, R.; Bucci, L.; Pini, E.; Nikkïla, J.; Monti, D.; Satokari, R.; Franceschi, C.; et al. Through Ageing, and Beyond: Gut Microbiota and Inflammatory Status in Seniors and Centenarians. PLoS ONE 2010, 5, e10667. [Google Scholar] [CrossRef]
- Jeffery, I.B.; Lynch, D.B.; O’Toole, P.W. Composition and Temporal Stability of the Gut Microbiota in Older Persons. ISME J. 2016, 10, 170–182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alkanani, A.K.; Hara, N.; Gottlieb, P.A.; Ir, D.; Robertson, C.E.; Wagner, B.D.; Frank, D.N.; Zipris, D. Alterations in Intestinal Microbiota Correlate With Susceptibility to Type 1 Diabetes. Diabetes 2015, 64, 3510–3520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clarke, S.F.; Murphy, E.F.; O’Sullivan, O.; Lucey, A.J.; Humphreys, M.; Hogan, A.; Hayes, P.; O’Reilly, M.; Jeffery, I.B.; Wood-Martin, R.; et al. Exercise and Associated Dietary Extremes Impact on Gut Microbial Diversity. Gut 2014, 63, 1913–1920. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carding, S.; Verbeke, K.; Vipond, D.T.; Corfe, B.M.; Owen, L.J. Dysbiosis of the Gut Microbiota in Disease. Microb. Ecol. Health Dis. 2015, 26. [Google Scholar] [CrossRef] [PubMed]
- Pomié, C.; Blasco-Baque, V.; Klopp, P.; Nicolas, S.; Waget, A.; Loubières, P.; Azalbert, V.; Puel, A.; Lopez, F.; Dray, C.; et al. Triggering the Adaptive Immune System with Commensal Gut Bacteria Protects against Insulin Resistance and Dysglycemia. Mol. Metab. 2016, 5, 392–403. [Google Scholar] [CrossRef] [PubMed]
- Morales, P.; Fujio, S.; Navarrete, P.; Ugalde, J.A.; Magne, F.; Carrasco-Pozo, C.; Tralma, K.; Quezada, M.; Hurtado, C.; Covarrubias, N.; et al. Impact of Dietary Lipids on Colonic Function and Microbiota: An Experimental Approach Involving Orlistat-Induced Fat Malabsorption in Human Volunteers. Clin. Transl. Gastroenterol. 2016, 7, e161. [Google Scholar] [CrossRef]
- Endesfelder, D.; Zu Castell, W.; Ardissone, A.; Davis-Richardson, A.G.; Achenbach, P.; Hagen, M.; Pflueger, M.; Gano, K.A.; Fagen, J.R.; Drew, J.C.; et al. Compromised Gut Microbiota Networks in Children with Anti-Islet Cell Autoimmunity. Diabetes 2014, 63, 2006–2014. [Google Scholar] [CrossRef] [Green Version]
- Ghosh, S.S.; Wang, J.; Yannie, P.; Ghosh, S. Intestinal Barrier Dysfunction, LPS Translocation, and Disease Development. J. Endocr. Soc. 2020, 4, bvz039. [Google Scholar] [CrossRef] [Green Version]
- Cani, P.D.; Bibiloni, R.; Knauf, C.; Waget, A.; Neyrinck, A.M.; Delzenne, N.M.; Burcelin, R. Changes in Gut Microbiota Control Metabolic Endotoxemia-Induced Inflammation in High-Fat Diet-Induced Obesity and Diabetes in Mice. Diabetes 2008, 57, 1470–1481. [Google Scholar] [CrossRef] [Green Version]
- Topchiy, E.; Cirstea, M.; Kong, H.; Boyd, J.; Wang, Y.; Russell, J.; Waley, K. Lipopolysaccharide Is Cleared from the Circulation by Hepatocytes via the Low Density Lipoprotein Receptor. PLoS ONE 2016, 11, e0155030. [Google Scholar] [CrossRef] [Green Version]
- Rohr, M.W.; Narasimhulu, C.; Rudeski-Rohr, T.; Parthasarathy, S. Negative Effects of a High-Fat Diet on Intestinal Permeability: A Review. Adv. Nutr. 2020, 11, 77–91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lam, Y.Y.; Ha, C.W.Y.; Hoffmann, J.M.A.; Oscarsson, J.; Dinudom, A.; Mather, T.J.; Cook, D.I.; Hunt, N.H.; Caterson, I.D.; Holmes, A.J.; et al. Effects of Dietary Fat Profile on Gut Permeability and Microbiota and Their Relationships with Metabolic Changes in Mice. Obesity 2015, 23, 1429–1439. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Al-Sadi, R.; Said, H.M.; Ma, T.Y. Lipopolysaccharide Causes an Increase in Intestinal Tight Junction Permeability in Vitro and in Vivo by Inducing Enterocyte Membrane Expression and Localization of TLR-4 and CD14. Am. J. Pathol. 2013, 182, 375–387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, S.; Nighot, M.; Al-Sadi, R.; Alhmoud, T.; Nighot, P.; Ma, T.Y. Lipopolysaccharide Regulation of Intestinal Tight Junction Permeability Is Mediated by TLR4 Signal Transduction Pathway Activation of FAK and MyD88. J. Immunol. 2015, 195, 4999–5010. [Google Scholar] [CrossRef]
- Garidou, L.; Pomié, C.; Klopp, P.; Waget, A.; Charpentier, J.; Aloulou, M.; Giry, A.; Serino, M.; Stenman, L.; Lahtinen, S.; et al. The Gut Microbiota Regulates Intestinal CD4 T Cells Expressing RORγt and Controls Metabolic Disease. Cell. Metab. 2015, 22, 100–112. [Google Scholar] [CrossRef] [Green Version]
- Cavallari, J.F.; Denou, E.; Foley, K.P.; Khan, W.I.; Schertzer, J.D. Different Th17 Immunity in Gut, Liver, and Adipose Tissues during Obesity: The Role of Diet, Genetics, and Microbes. Gut Microbes 2016, 7, 82–89. [Google Scholar] [CrossRef] [Green Version]
- Gomes, J.; de Assis, J.; Gonçalves, R. Metabolic endotoxemia and diabetes mellitus: A systematic review. Metabolism 2017, 86, 133–144. [Google Scholar] [CrossRef]
- Matheus, V.A.; Monteiro, L.; Oliveira, R.B.; Maschio, D.A.; Collares-Buzato, C.B. Butyrate Reduces High-Fat Diet-Induced Metabolic Alterations, Hepatic Steatosis and Pancreatic Beta Cell and Intestinal Barrier Dysfunctions in Prediabetic Mice. Exp. Biol. Med. 2017. [Google Scholar] [CrossRef] [Green Version]
- Pedersen, C.; Gallagher, E.; Horton, F.; Ellis, R.J.; Ijaz, U.Z.; Wu, H.; Jaiyeola, E.; Diribe, O.; Duparc, T.; Cani, P.D.; et al. Host–Microbiome Interactions in Human Type 2 Diabetes Following Prebiotic Fibre (Galacto-Oligosaccharide) Intake. Br. J. Nutr. 2016, 116, 1869–1877. [Google Scholar] [CrossRef] [Green Version]
- Song, M.J.; Kim, K.H.; Yoon, J.M.; Kim, J.B. Activation of Toll-like Receptor 4 Is Associated with Insulin Resistance in Adipocytes. Biochem. Biophys. Res. Commun. 2006, 346, 739–745. [Google Scholar] [CrossRef]
- Amyot, J.; Semache, M.; Ferdaoussi, M.; Fontés, G.; Poitout, V. Lipopolysaccharides Impair Insulin Gene Expression in Isolated Islets of Langerhans via Toll-Like Receptor-4 and NF-ΚB Signalling. PLoS ONE 2012, 7, e36200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, J.; Peng, J.; An, H.; He, Q.; Boronina, T.; Guo, S.; White, M.F.; Cole, P.A.; He, L. Endotoxemia-Mediated Activation of Acetyltransferase P300 Impairs Insulin Signaling in Obesity. Nat. Commun. 2017, 8, 131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kelder, T.; Stroeve, J.H.M.; Bijlsma, S.; Radonjic, M.; Roeselers, G. Correlation Network Analysis Reveals Relationships between Diet-Induced Changes in Human Gut Microbiota and Metabolic Health. Nutr. Diabetes 2014, 4, e122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mariat, D.; Firmesse, O.; Levenez, F.; Guimarăes, V.; Sokol, H.; Doré, J.; Corthier, G.; Furet, J.P. The Firmicutes/Bacteroidetes ratio of the human microbiota changes with age. BMC Microbiol. 2009, 9, 123. [Google Scholar] [CrossRef]
- Sikalidis, A.; Maykish, A. The Gut Microbiome and Type 2 Diabetes Mellitus: Discussing a Complex Relationship. Biomedicines 2020, 8, 8. [Google Scholar] [CrossRef] [Green Version]
- Magne, F.; Gotteland, M.; Gauthier, L.; Zazueta, A.; Pesoa, S.; Navarrete, P.; Balamurugan, R. The Firmicutes/Bacteroidetes Ratio: A Relevant Marker of Gut Dysbiosis in Obese Patients. Nutrients 2020, 12, 1474. [Google Scholar] [CrossRef]
- Larsen, N.; Vogensen, F.; Van den Berg, F.; Nielsen, D.S.; Andreasen, A.S.; Pedersen, B.K.; Al-Soud, W.A.; Sørensen, S.J.; Hansen, L.H.; Jakobsen, M. Gut Microbiota in Human Adults with Type 2 Diabetes Differs from Non-Diabetic Adults. PLoS ONE 2010, 5, e9085. [Google Scholar] [CrossRef]
- Endesfelder, D.; Engel, M.; Davis-Richardson, A.G.; Ardissone, A.N.; Achenbach, P.; Hummel, S.; Winkler, C.; Atkinson, M.; Schatz, D.; Triplett, E.; et al. Towards a Functional Hypothesis Relating Anti-Islet Cell Autoimmunity to the Dietary Impact on Microbial Communities and Butyrate Production. Microbiome 2016, 4, 17. [Google Scholar] [CrossRef] [Green Version]
- Ruiz, L.; Delgado, S.; Ruas-Madiedo, P.; Sánchez, B.; Margolles, A. Bifidobacteria and Their Molecular Communication with the Immune System. Front. Microbiol. 2017, 8, 2345. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.; Lian, F.; Zhao, L.; Zhao, Y.; Chen, X.; Zhang, X.; Guo, Y.; Zhang, C.; Zgou, Q.; Xue, Z.; et al. Structural modulation of gut microbiota during alleviation of type 2 diabetes with a Chinese herbal formula. ISME J. 2015, 9, 552–562. [Google Scholar] [CrossRef]
- Yanagibashi, T.; Hosono, A.; Oyama, A.; Tsuda, M.; Suzuki, A.; Hachimura, S.; Takahashi, Y.; Momose, Y.; Itoh, K.; Hirayama, K.; et al. IgA Production in the Large Intestine Is Modulated by a Different Mechanism than in the Small Intestine: Bacteroides Acidifaciens Promotes IgA Production in the Large Intestine by Inducing Germinal Center Formation and Increasing the Number of IgA+ B Cells. Immunobiology 2013, 218, 645–651. [Google Scholar] [CrossRef] [PubMed]
- Murri, M.; Leiva, I.; Gomez-Zumaquero, J.M.; Tinahones, F.J.; Cardona, F.; Soriguer, F.; Queipo-Ortuño, M.I. Gut Microbiota in Children with Type 1 Diabetes Differs from That in Healthy Children: A Case-Control Study. BMC Med. 2013, 11, 46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jandhyala, S.M.; Madhulika, A.; Deepika, G.; Rao, G.V.; Reddy, D.N.; Subramanyam, C.; Sasikala, M.; Talukdar, R. Altered Intestinal Microbiota in Patients with Chronic Pancreatitis: Implications in Diabetes and Metabolic Abnormalities. Sci. Rep. 2017, 7, 43640. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Remely, M.; Aumueller, E.; Merold, C.; Dworzak, S.; Hippe, B.; Zanner, J.; Pointner, A.; Brath, H.; Haslberger, A.G. Effects of Short Chain Fatty Acid Producing Bacteria on Epigenetic Regulation of FFAR3 in Type 2 Diabetes and Obesity. Gene 2014, 537, 85–92. [Google Scholar] [CrossRef]
- Gurung, M.; Li, Z.; You, H.; Rodrigues, R.; Jump, D.B.; Morgun, A.; Shulzhenko, N. Role of gut microbiota in type 2 diabetes pathophysiology. EBioMedicine 2020, 51, 102590. [Google Scholar] [CrossRef] [Green Version]
- Kim, C.H. Microbiota or Short-Chain Fatty Acids: Which Regulates Diabetes. Cell. Mol. Immunol. 2018, 15, 88–91. [Google Scholar] [CrossRef]
- García, P.O. La fibra alimentaria y su uso terapéutico en algunas enfermedades crónicas. Gac. Med. Caracas 2020, 120, 107–114. [Google Scholar]
- Puddu, A.; Sanguineti, R.; Montecucco, F.; Viviani, G.L. Evidence for the Gut Microbiota Short-Chain Fatty Acids as Key Pathophysiological Molecules Improving Diabetes. Mediat. Inflamm. 2014, 2014, 162021. [Google Scholar] [CrossRef]
- Myhrstad, M.C.W.; Tunsjø, H.; Charnock, C.; Telle-Hansen, V.H. Dietary Fiber, Gut Microbiota, and Metabolic Regulation-Current Status in Human Randomized Trials. Nutrients 2020, 12, 859. [Google Scholar] [CrossRef] [Green Version]
- Ratajczak, W.; Rył, A.; Mizerski, A.; Walczakiewicz, K.; Sipak, O.; Laszczyńska, M. Immunomodulatory Potential of Gut Microbiome-Derived Short-Chain Fatty Acids (SCFAs). Acta Biochim. Pol. 2019, 66, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Dávila, L.A.; Pirela, V.B.; Díaz, W.; Villasmil, N.R.; León, S.C.; Contreras, M.C.E.; Bonacich, K.B.; Agüero, S.D.; Vergara, P.C.; Bonacich, R.B.; et al. The Microbiome and the Epigenetics of Diabetes Mellitus. In Diabetes Food Plan; Waisundara, V., Ed.; InTech: London, UK, 2018. [Google Scholar]
- Kimura, I.; Ichimura, A.; Ohue-Kitano, R.; Igarashi, M. Free Fatty Acid Receptors in Health and Disease. Physiol. Rev. 2020, 100, 171–210. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.U.; In, H.J.; Kwon, M.S.; Park, B.; Jo, M.; Kim, M.-O.; Cho, S.; Lee, S.; Lee, H.-J.; Kwak, Y.S.; et al. β-Arrestin 2 Mediates G Protein-Coupled Receptor 43 Signals to Nuclear Factor-ΚB. Biol. Pharm. Bull. 2013, 36, 1754–1759. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cox, M.A.; Jackson, J.; Stanton, M.; Rojas-Triana, A.; Bober, L.; Laverty, M.; Yang, X.; Zhu, F.; Liu, J.; Wang, S.; et al. Short-Chain Fatty Acids Act as Antiinflammatory Mediators by Regulating Prostaglandin E(2) and Cytokines. World J. Gastroenterol. 2009, 15, 5549–5557. [Google Scholar] [CrossRef] [PubMed]
- Hernández, M.A.G.; Canfora, E.E.; Jocken, J.W.E.; Blaak, E.E. The Short-Chain Fatty Acid Acetate in Body Weight Control and Insulin Sensitivity. Nutrients 2019, 11, 1943. [Google Scholar] [CrossRef] [Green Version]
- Larasati, R.A.; Harbuwono, D.S.; Rahajeng, E.; Pradipta, S.; Nuraeni, H.S.; Susilowati, A.; Wibowo, H. The Role of Butyrate on Monocyte Migration and Inflammation Response in Patient with Type 2 Diabetes Mellitus. Biomedicines 2019, 7, 74. [Google Scholar] [CrossRef] [Green Version]
- Chun, E.; Lavoie, S.; Fonseca-Pereira, D.; Bae, S.; Michaud, M.; Hoveyda, H.R.; Fraser, G.L.; Gallini Comeau, C.A.; Glickman, J.N.; Fuller, M.H.; et al. Metabolite-Sensing Receptor Ffar2 Regulates Colonic Group 3 Innate Lymphoid Cells and Gut Immunity. Immunity 2019, 51, 871–884. [Google Scholar] [CrossRef]
- Wu, W.; Sun, M.; Chen, F.; Cao, A.T.; Liu, H.; Zhao, Y.; Huang, X.; Xiao, Y.; Yao, S.; Zhao, Q.; et al. Microbiota Metabolite Short-Chain Fatty Acid Acetate Promotes Intestinal IgA Response to Microbiota Which Is Mediated by GPR43. Mucosal. Immunol. 2017, 10, 946–956. [Google Scholar] [CrossRef] [Green Version]
- Kim, C.H.; Park, J.; Kim, M. Gut Microbiota-Derived Short-Chain Fatty Acids, T Cells, and Inflammation. Immune Netw. 2014, 14, 277–288. [Google Scholar] [CrossRef] [Green Version]
- Astakhova, L.; Ngara, M.; Babich, O.; Prosekov, A.; Asyakina, L.; Dyshlyuk, L.; Midtvedt, T.; Zhou, X.; Ernberg, I.; Matskova, L. Short Chain Fatty Acids (SCFA) Reprogram Gene Expression in Human Malignant Epithelial and Lymphoid Cells. PLoS ONE 2016, 11, e0154102. [Google Scholar] [CrossRef]
- Yap, Y.A.; Mariño, E. Dietary SCFAs Immunotherapy: Reshaping the Gut Microbiota in Diabetes. In SpringerLink; Springer: New York, NY, USA, 2020. [Google Scholar] [CrossRef]
- Mariño, E.; Richards, J.L.; McLeod, K.H.; Stanley, D.; Yap, Y.A.; Knight, J.; McKenzie, C.; Kranich, J.; Oliveira, A.C.; Rossello, F.J.; et al. Gut Microbial Metabolites Limit the Frequency of Autoimmune T Cells and Protect against Type 1 Diabetes. Nat. Immunol. 2017, 18, 552–562. [Google Scholar] [CrossRef]
- Carpio Duran, A.L.; Duran Medina, M.F.; Andrade Valdivieso, M.R.; Espinoza Dunn, M.A.; Rodas Torres, W.P.; Abad Barrera, L.N.; Rodríguez Barzola, C.V.; Yagual Villon, O.A. Terapia Incretinomimética: Evidencia Clínica de La Eficacia de Los Agonistas Del GLP-1R y Sus Efectos Cardio-Protectores. Rev. Latinoam. Hipertens. 2018, 13, 400–415. [Google Scholar]
- Rahat-Rozenbloom, S.; Fernandes, J.; Cheng, J.; Wolever, T.M.S. Acute Increases in Serum Colonic Short-Chain Fatty Acids Elicited by Inulin Do Not Increase GLP-1 or PYY Responses but May Reduce Ghrelin in Lean and Overweight Humans. Eur. J. Clin. Nutr. 2017, 71, 953–958. [Google Scholar] [CrossRef] [PubMed]
- Bjerg, A.T.; Kristensen, M.; Ritz, C.; Holst, J.J.; Rasmussen, C.; Leser, T.D.; Wellejus, A.; Astrup, A. Lactobacillus Paracasei Subsp Paracasei L. Casei W8 Suppresses Energy Intake Acutely. Appetite 2014, 82, 111–118. [Google Scholar] [CrossRef] [PubMed]
- De Velasco, P.; Ferreira, A.; Crovesy, L.; Marine, T.; Das Graças Tavares do Carmo, M. Fatty Acids, Gut Microbiota, and the Genesis of Obesity. In Biochemistry and Health Benefits of Fatty Acids; Waisundara, V., Ed.; IntechOpen: London, UK, 2018. [Google Scholar]
- Grasset, E.; Puel, A.; Charpentier, J.; Collet, X.; Christensen, J.E.; Tercé, F.; Burcelin, R. A Specific Gut Microbiota Dysbiosis of Type 2 Diabetic Mice Induces GLP-1 Resistance through an Enteric NO-Dependent and Gut-Brain Axis Mechanism. Cell Metab. 2017, 25, 1075–1090. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perry, R.J.; Peng, L.; Barry, N.A.; Cline, G.W.; Zhang, D.; Cardone, R.L.; Petersen, K.F.; Kibbey, R.G.; Goodman, A.L.; Shulman, G.I. Acetate Mediates a Microbiome–Brain–β-Cell Axis to Promote Metabolic Syndrome. Nature 2016, 534, 213–217. [Google Scholar] [CrossRef] [Green Version]
- Yang, M.; Wang, J.; Wu, S.; Yuan, L.; Zhao, X.; Liu, C.; Xie, J.; Jia, Y.; Lai, Y.; Zhao, A.Z.; et al. Duodenal GLP-1 Signaling Regulates Hepatic Glucose Production through a PKC-δ-Dependent Neurocircuitry. Cell Death Dis. 2017, 8, e2609. [Google Scholar] [CrossRef] [Green Version]
- Vieira, A.T.; Fukumori, C.; Ferreira, C.M. New Insights into Therapeutic Strategies for Gut Microbiota Modulation in Inflammatory Diseases. Clin. Transl. Immunol. 2016, 5, e87. [Google Scholar] [CrossRef]
- Kellow, N.J.; Coughlan, M.T.; Savige, G.S.; Reid, C.M. Effect of Dietary Prebiotic Supplementation on Advanced Glycation, Insulin Resistance and Inflammatory Biomarkers in Adults with Pre-Diabetes: A Study Protocol for a Double-Blind Placebo-Controlled Randomised Crossover Clinical Trial. BMC Endocr. Disord. 2014, 14, 55. [Google Scholar] [CrossRef] [Green Version]
- Kassaian, N.; Aminorroaya, A.; Feizi, A.; Jafari, P.; Amini, M. The Effects of Probiotic and Synbiotic Supplementation on Metabolic Syndrome Indices in Adults at Risk of Type 2 Diabetes: Study Protocol for a Randomized Controlled Trial. Trials 2017, 18, 148. [Google Scholar] [CrossRef] [Green Version]
- Dávila, L.A.; Pirela, V.B.; Villasmil, N.R.; Cisternas, S.; Díaz, W.; Escobar, M.C.; Carrasco, P.; Durán, S.; Buhring, K.; Buhring, R.; et al. New Insights into Alleviating Diabetes Mellitus: Role of Gut Microbiota and a Nutrigenomic Approac. In Diabetes Food Plan; Waisundara, V., Ed.; InTech: London, UK, 2018. [Google Scholar]
- Bolívar González, S.; Talero Barrientos, E.; Motilva Sánchez, V. Efectos de Un Preparado Probiótico En Un Modelo de Colitis Experimental Crónica En Ratones, Inducida Por La Ingesta de Dextrano Sulfato Sódico (DSS). Cienc. Innov. Salud 2015, 3. [Google Scholar] [CrossRef]
- Yao, K.; Zeng, L.; He, Q.; Wang, W.; Lei, J.; Zou, X. Effect of Probiotics on Glucose and Lipid Metabolism in Type 2 Diabetes Mellitus: A Meta-Analysis of 12 Randomized Controlled Trials. Med. Sci. Monit. 2017, 23, 3044–3053. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Naito, E.; Yoshida, Y.; Makino, K.; Kounoshi, Y.; Kunihiro, S.; Takahashi, R.; Matsuzaki, T.; Miyazaki, K.; Ishikawa, F. Beneficial Effect of Oral Administration of Lactobacillus Casei Strain Shirota on Insulin Resistance in Diet-Induced Obesity Mice. J. Appl. Microbiol. 2011, 110, 650–657. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.J.; Wang, R.; Li, X.; Wang, R. Bifidobacterium Longum Supplementation Improved High-Fat-Fed-Induced Metabolic Syndrome and Promoted Intestinal Reg I Gene Expression. Exp. Biol. Med. 2011, 236, 823–831. [Google Scholar] [CrossRef] [PubMed]
- Alfa, M.J.; Strang, D.; Tappia, P.S.; Olson, N.; De Gagne, P.; Bray, D.; Murray, B.-L.; Hiebert, B. A Randomized Placebo Controlled Clinical Trial to Determine the Impact of Digestion Resistant Starch MSPrebiotic® on Glucose, Insulin, and Insulin Resistance in Elderly and Mid-Age Adults. Front. Med. 2017, 4, 260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Vadder, F.; Kovatcheva-Datchary, P.; Goncalves, D.; Vinera, J.; Zitoun, C.; Duchampt, A.; Bäckhed, F.; Mithieux, G. Microbiota-Generated Metabolites Promote Metabolic Benefits via Gut-Brain Neural Circuits. Cell 2014, 156, 84–96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, J.; Yuan, X.; Cheng, G.; Jiao, S.; Feng, C.; Zhao, X.; Yin, H.; Du, Y.; Liu, H. Chitosan Oligosaccharides Improve the Disturbance in Glucose Metabolism and Reverse the Dysbiosis of Gut Microbiota in Diabetic Mice. Carbohydr. Polym. 2018, 190, 77–86. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.; Hyslop, C.M.; Shrivastava, V.; Ochoa, A.; Reimer, R.A.; Huang, C. Oligofructose as an Adjunct in Treatment of Diabetes in NOD Mice. Sci. Rep. 2016, 6, 37627. [Google Scholar] [CrossRef] [Green Version]
- Everard, A.; Matamoros, S.; Geurts, L.; Delzenne, N.M.; Cani, P.D. Saccharomyces Boulardii Administration Changes Gut Microbiota and Reduces Hepatic Steatosis, Low-Grade Inflammation, and Fat Mass in Obese and Type 2 Diabetic Db/Db Mice. MBio 2014, 5, e01011. [Google Scholar] [CrossRef] [Green Version]
- Karczewski, J.; Troost, F.J.; Konings, I.; Dekker, J.; Kleerebezem, M.; Brummer, R.-J.M.; Wells, J.M. Regulation of Human Epithelial Tight Junction Proteins by Lactobacillus Plantarum in Vivo and Protective Effects on the Epithelial Barrier. Am. J. Physiol. Gastrointest. Liver Physiol. 2010, 298, G851–G859. [Google Scholar] [CrossRef] [Green Version]
- Sharma, P.; Bhardwaj, P.; Singh, R. Administration of Lactobacillus Casei and Bifidobacterium Bifidum Ameliorated Hyperglycemia, Dyslipidemia, and Oxidative Stress in Diabetic Rats. Int. J. Prev. Med. 2016, 7. [Google Scholar] [CrossRef]
- Valladares, R.; Sankar, D.; Li, N.; Williams, E.; Lai, K.-K.; Abdelgeliel, A.S.; Gonzalez, C.F.; Wasserfall, C.H.; Iii, J.L.; Schatz, D.; et al. Lactobacillus Johnsonii N6.2 Mitigates the Development of Type 1 Diabetes in BB-DP Rats. PLoS ONE 2010, 5, e10507. [Google Scholar] [CrossRef] [PubMed]
- Balakumar, M.; Prabhu, D.; Sathishkumar, C.; Prabu, P.; Rokana, N.; Kumar, R.; Raghavan, S.; Soundarajan, A.; Grover, S.; Batish, V.K.; et al. Improvement in Glucose Tolerance and Insulin Sensitivity by Probiotic Strains of Indian Gut Origin in High-Fat Diet-Fed C57BL/6J Mice. Eur. J. Nutr. 2016, 57, 279–295. [Google Scholar] [CrossRef] [PubMed]
- Yadav, H.; Lee, J.-H.; Lloyd, J.; Walter, P.; Rane, S.G. Beneficial Metabolic Effects of a Probiotic via Butyrate-Induced GLP-1 Hormone Secretion. J. Biol. Chem. 2013, 288, 25088–25097. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asemi, Z.; Khorrami-Rad, A.; Alizadeh, S.-A.; Shakeri, H.; Esmaillzadeh, A. Effects of Synbiotic Food Consumption on Metabolic Status of Diabetic Patients: A Double-Blind Randomized Cross-over Controlled Clinical Trial. Clin. Nutr. 2014, 33, 198–203. [Google Scholar] [CrossRef] [PubMed]
- Ebrahimi, Z.S.; Nasli-Esfahani, E.; Nadjarzade, A.; Mozaffari-Khosravi, H. Effect of Symbiotic Supplementation on Glycemic Control, Lipid Profiles and Microalbuminuria in Patients with Non-Obese Type 2 Diabetes: A Randomized, Double-Blind, Clinical Trial. J. Diabetes Metab. Disord. 2017, 16, 23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, T.-C.D. Diet and Gut Microbiota in Health and Disease. In Intestinal Microbiome: Functional Aspects in Health and Disease; Nestlé Nutrition Institute Workshop Series; Karger Publishers: Basel, Switzerland, 2017; Volume 88, pp. 117–126. [Google Scholar] [CrossRef] [Green Version]
- Heianza, Y.; Sun, D.; Li, X.; DiDonato, J.A.; Bray, G.A.; Sacks, F.M.; Qi, L. Gut Microbiota Metabolites, Amino Acid Metabolites and Improvements in Insulin Sensitivity and Glucose Metabolism: The POUNDS Lost Trial. Gut 2018, 67. [Google Scholar] [CrossRef]
- Miranda, P.J.P.; Cueva, C. Rol de la metformina en el tratamiento de la diabetes mellitus gestacional: Situación actual. Arch. Venez. Farmacol. Ter. 2019, 38, 234–239. [Google Scholar]
- Napolitano, A.; Miller, S.; Nicholls, A.W.; Baker, D.; Van Horn, S.; Thomas, E.; Rajpal, D.; Spivak, A.; Brown, J.R.; Nunez, D.J. Novel Gut-Based Pharmacology of Metformin in Patients with Type 2 Diabetes Mellitus. PLoS ONE 2014, 9, e100778. [Google Scholar] [CrossRef]
- Bonora, E.; Cigolini, M.; Bosello, O.; Zancanaro, C.; Capretti, L.; Zavaroni, I.; Coscelli, C.; Butturini, U. Lack of Effect of Intravenous Metformin on Plasma Concentrations of Glucose, Insulin, C-Peptide, Glucagon and Growth Hormone in Non-Diabetic Subjects. Curr. Med. Res. Opin. 1984, 9, 47–51. [Google Scholar] [CrossRef]
- Ridlon, J.M.; Harris, S.C.; Bhowmik, S.; Kang, D.-J.; Hylemon, P.B. Consequences of Bile Salt Biotransformations by Intestinal Bacteria. Gut Microbes 2016, 7, 22–39. [Google Scholar] [CrossRef] [Green Version]
- Forslund, K.; Hildebrand, F.; Nielsen, T.; Falony, G.; Le Chatelier, E.; Sunagawa, S.; Prifti, E.; Vieira-Silva, S.; Gudmundsdottir, V.; Krogh Pedersen, H.; et al. Disentangling Type 2 Diabetes and Metformin Treatment Signatures in the Human Gut Microbiota. Nature 2015, 528, 262–266. [Google Scholar] [CrossRef] [PubMed]
- De La Cuesta-Zuluaga, J.; Mueller, N.T.; Corrales-Agudelo, V.; Velásquez-Mejía, E.P.; Carmona, J.A.; Abad, J.M.; Escobar, J.S. Metformin Is Associated With Higher Relative Abundance of Mucin-Degrading Akkermansia Muciniphila and Several Short-Chain Fatty Acid–Producing Microbiota in the Gut. Diabetes Care 2016, dc161324. [Google Scholar] [CrossRef] [Green Version]
- Wu, H.; Esteve, E.; Tremaroli, V.; Khan, M.T.; Caesar, R.; Mannerås-Holm, L.; Ståhlman, M.; Olsson, L.M.; Serino, M.; Planas-Fèlix, M.; et al. Metformin Alters the Gut Microbiome of Individuals with Treatment-Naive Type 2 Diabetes, Contributing to the Therapeutic Effects of the Drug. Nat. Med. 2017, 23, 850–858. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Ko, G. Effect of Metformin on Metabolic Improvement and Gut Microbiota. Appl. Environ. Microbiol. 2014, 80, 5935–5943. [Google Scholar] [CrossRef] [Green Version]
- Vallianou, N.; Stratigou, T.; Tsagarakis, S. Metformin and gut microbiota: their interactions and their impact on diabetes. Hormones (Athens) 2019, 2, 141–144. [Google Scholar] [CrossRef]
- Bryrup, T.; Thomsen, C.W.; Kern, T.; Allin, K.H.; Brandslund, I.; Jørgensen, N.R.; Vestergaard, H.; Hansen, T.; Hansen, T.H.; Pedersen, O.; et al. Metformin-Induced Changes of the Gut Microbiota in Healthy Young Men: Results of a Non-Blinded, One-Armed Intervention Study. Diabetologia 2019, 62, 1024–1035. [Google Scholar] [CrossRef] [Green Version]
- Su, B.; Liu, H.; Li, J.; Sunli, Y.; Liu, B.; Liu, D.; Zhang, P.; Meng, X. Acarbose Treatment Affects the Serum Levels of Inflammatory Cytokines and the Gut Content of Bifidobacteria in Chinese Patients with Type 2 Diabetes Mellitus. J. Diabetes 2015, 7, 729–739. [Google Scholar] [CrossRef]
- Gu, Y.; Wang, X.; Li, J.; Zhang, Y.; Zhong, H.; Liu, R.; Zhang, D.; Feng, Q.; Xie, X.; Hong, J.; et al. Analyses of Gut Microbiota and Plasma Bile Acids Enable Stratification of Patients for Antidiabetic Treatment. Nat. Commun. 2017, 8, 1785. [Google Scholar] [CrossRef]
- Smith, B.J.; Miller, R.A.; Ericsson, A.C.; Harrison, D.C.; Strong, R.; Schmidt, T.M. Changes in the Gut Microbiome and Fermentation Products Concurrent with Enhanced Longevity in Acarbose-Treated Mice. BMC Microbiol. 2019, 19, 130. [Google Scholar] [CrossRef] [Green Version]
- Baxter, N.T.; Lesniak, N.A.; Sinani, H.; Schloss, P.D.; Koropatkin, N.M. The Glucoamylase Inhibitor Acarbose Has a Diet-Dependent and Reversible Effect on the Murine Gut Microbiome. MSphere 2019, 4. [Google Scholar] [CrossRef] [Green Version]
- Zhao, L.; Chen, Y.; Xia, F.; Abudukerimu, B.; Zhang, W.; Guo, Y.; Wang, N.; Lu, Y. A Glucagon-Like Peptide-1 Receptor Agonist Lowers Weight by Modulating the Structure of Gut Microbiota. Front. Endocrinol. 2018, 9, 233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Li, P.; Tang, Z.; Yan, X.; Feng, B. Structural Modulation of the Gut Microbiota and the Relationship with Body Weight: Compared Evaluation of Liraglutide and Saxagliptin Treatment. Sci. Rep. 2016, 6, 33251. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Saha, S.; Van Horn, S.; Thomas, E.; Traini, C.; Sathe, G.; Rajpal, D.K.; Brown, J.R. Gut Microbiome Differences between Metformin- and Liraglutide-Treated T2DM Subjects. Endocrinol. Diabetes Metab. 2018, 1, e00009. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moreira, G.V.; Azevedo, F.F.; Ribeiro, L.M.; Santos, A.; Guadagnini, D.; Gama, P.; Liberti, E.A.; Saad, M.; Carvalho, C. Liraglutide Modulates Gut Microbiota and Reduces NAFLD in Obese Mice. J. Nutr. Biochem. 2018, 62, 143–154. [Google Scholar] [CrossRef]
- Sun, L.; Xie, C.; Wang, G.; Wu, Y.; Wu, Q.; Wang, X.; Liu, J.; Deng, Y.; Xia, J.; Chen, B.; et al. Gut Microbiota and Intestinal FXR Mediate the Clinical Benefits of Metformin. Nat. Med. 2018, 24, 1919–1929. [Google Scholar] [CrossRef]
- Fang, S.; Suh, J.M.; Reilly, S.M.; Yu, E.; Osborn, O.; Lackey, D.; Yoshihara, E.; Perino, A.; Jacinto, S.; Lukasheva, Y.; et al. Intestinal FXR Agonism Promotes Adipose Tissue Browning and Reduces Obesity and Insulin Resistance. Nat. Med. 2015, 21, 159–165. [Google Scholar] [CrossRef] [Green Version]
- Pathak, P.; Xie, C.; Nichols, R.G.; Ferrell, J.M.; Boehme, S.; Krausz, K.W.; Patterson, A.D.; Gonzalez, F.J.; Chiang, J.Y.L. Intestine Farnesoid X Receptor Agonist and the Gut Microbiota Activate G-Protein Bile Acid Receptor-1 Signaling to Improve Metabolism. Hepatology 2018, 68, 1574–1588. [Google Scholar] [CrossRef]
- Zhang, X.; Fang, Z.; Zhang, C.; Xia, H.; Jie, Z.; Han, X.; Chen, Y.; Ji, L. Effects of Acarbose on the Gut Microbiota of Prediabetic Patients: A Randomized, Double-Blind, Controlled Crossover Trial. Diabetes Ther. 2017, 8, 293–307. [Google Scholar] [CrossRef] [Green Version]
- Remely, M.; Hippe, B.; Zanner, J.; Aumueller, E.; Brath, H.; Haslberger, A.G. Gut Microbiota of Obese, Type 2 Diabetic Individuals Is Enriched in Faecalibacterium Prausnitzii, Akkermansia Muciniphila and Peptostreptococcus Anaerobius after Weight Loss. Endocr. Metab. Immune Disord. Drug Targets 2016, 16, 99–106. [Google Scholar] [CrossRef]
- Xu, G.-D.; Cai, L.; Ni, Y.-S.; Tian, S.-Y.; Lu, Y.-Q.; Wang, L.-N.; Chen, L.-L.; Ma, W.-Y.; Deng, S.-P. Comparisons of Effects on Intestinal Short-Chain Fatty Acid Concentration after Exposure of Two Glycosidase Inhibitors in Mice. Biol. Pharm. Bull. 2018, 41, 1024–1033. [Google Scholar] [CrossRef]
- Zhang, Q.; Xiao, X.; Zheng, J.; Li, M.; Yu, M.; Ping, F.; Wang, T.; Wang, X. Featured Article: Structure Moderation of Gut Microbiota in Liraglutide-Treated Diabetic Male Rats. Exp. Biol. Med. 2018, 243, 34–44. [Google Scholar] [CrossRef] [PubMed]



| Therapeutic Strategy | Molecule/ Microorganism | Subject of Study | Effects | Ref. |
|---|---|---|---|---|
| Prebiotics | Chitosan oligosaccharides | Mice | ↓Glycemia, IR, inflammatory mediators, lipogenesis ↑Occludins, intestinal integrity ↑ Bacteroidetes and Akkermansia ↓ Firmicutes and Helicobacter | [113] |
| Oligofructose | Mice | ↑Insulin and sensitivity to it ↓Lymphocyte infiltration to pancreatic islets. ↑Bifidobacterium ↓Clostridium leptum | [114] | |
| Inulin/Oligofructose | Humans | ↓Intestinal permeability, oxidative stress, inflammation, IR, and hyperglycemia. ↑Weight loss ↑ Bifidobacterium and Lactobacillus | [104] | |
| Probiotics | Saccharomyces boulardii | Mice | ↓Weight and body mass; hepatic steatosis, and inflammatory state ↑ Bacteroidetes ↓Firmicutes, Proteobacteria, Tenericutes | [115] |
| Lactobacillus Plantarum | Humans | Activation of TLR-2 ↑Binding proteins and protective function of the epithelium | [116] | |
| Lactobacillus casei Bifidobacterium (alone or in combination) | Mice | ↓Fasting glucose ↓HbA1c (B. bifidum and in combination) ↑Blood insulin and muscle glycogen Changes to the lipid profile and antioxidant effects | [117] | |
| Lactobacillus johnsonii N6.21 | Mice | ↓DM incidence and oxidative stress ↑Binding proteins | [118] | |
| Lactobacillus fermentum | Mice | ↓IR, blood glucose, total cholesterol, TAG, adiponectin, intestinal permeability, pro-inflammatory cytokines, and ER stress. ↑GLP-1 | [119] | |
| Lactobacillus rhamnosus | ||||
| VSL#3 (Bifidobacterium, Lactobacillus y Streptococcus) | Mice | ↓Weight gain, TAG and FA levels, IR and hyperinsulinemia, hepatic steatosis, and proinflammatory cytokines. Modulation of intestinal microbiota ↑Butyrate and GLP-1 | [120] | |
| Synbiotics | Lactobacillus sporogenes Inulin, isomalt, sorbitol y Stevia | Humans | ↓insulin, glutathione, uric acid and PCR ↑HDL cholesterol | [121] |
| Lactobacillus, Bifidobacterium, Streptococcus thermophilus Fructooligosaccharides | Humans | ↓fasting glucose, HbA1c levels, and BMI | [122] |
| Therapeutic Strategy | Subjects of Study | Effects | Ref. |
|---|---|---|---|
| Metformin | Mice and humans | ↑ Propionate and butyrate ↓ Intestinibacter spp. and Clostridium spp. ↑ Escherichia/Shigella spp. ↑ Akkermansia muciniphila | [130,132,133,134] |
| Acarbose | Mice and humans | ↓ LPS and proinflammatory cytokines ↑ Propionate and butyrate ↓ Clostridium and Bacteroides ↑ Bifidobacterium and Lactobacillus | [135,136,137,138] |
| Liraglutide | Mice and humans | ↑ Bacteroidetes ↑ Akkermansia muciniphila ↓ Firmicutes and Proteobacteria | [139,140,141,142] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Salazar, J.; Angarita, L.; Morillo, V.; Navarro, C.; Martínez, M.S.; Chacín, M.; Torres, W.; Rajotia, A.; Rojas, M.; Cano, C.; et al. Microbiota and Diabetes Mellitus: Role of Lipid Mediators. Nutrients 2020, 12, 3039. https://0-doi-org.brum.beds.ac.uk/10.3390/nu12103039
Salazar J, Angarita L, Morillo V, Navarro C, Martínez MS, Chacín M, Torres W, Rajotia A, Rojas M, Cano C, et al. Microbiota and Diabetes Mellitus: Role of Lipid Mediators. Nutrients. 2020; 12(10):3039. https://0-doi-org.brum.beds.ac.uk/10.3390/nu12103039
Chicago/Turabian StyleSalazar, Juan, Lissé Angarita, Valery Morillo, Carla Navarro, María Sofía Martínez, Maricarmen Chacín, Wheeler Torres, Arush Rajotia, Milagros Rojas, Clímaco Cano, and et al. 2020. "Microbiota and Diabetes Mellitus: Role of Lipid Mediators" Nutrients 12, no. 10: 3039. https://0-doi-org.brum.beds.ac.uk/10.3390/nu12103039