Pelargonium sidoides Root Extract: Simultaneous HPLC Separation, Determination, and Validation of Selected Biomolecules and Evaluation of SARS-CoV-2 Inhibitory Activity
Abstract
:1. Introduction
2. Results and Discussion
2.1. HPLC Method Development and Optimization
2.2. Method Validation
2.2.1. Specificity
2.2.2. Linearity
2.2.3. Detection and Quantitation Limits
- S = Standard deviation of the response
- b = Slope of the calibration curve
2.2.4. Precision and Accuracy
2.2.5. Robustness
2.3. Determination of the Four Biomolecules in Kalobin®, Umca® Solutions and Umca® Tablets
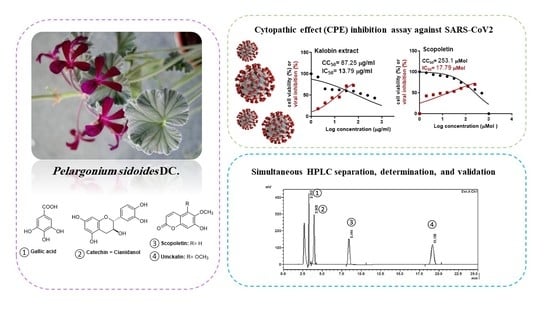
2.4. In Vitro Inhibitory Activity against SARS-CoV-2
2.5. In Silico Investigation of the Physicochemical and Pharmacokinetics Properties Using SwissADME Online Platform
2.6. Docking Study
3. Materials and Methods
3.1. Chromatographic Procedures
3.1.1. Solvents and Mobile Phases
3.1.2. Samples
3.1.3. Instrumentation and Chromatographic Conditions
3.1.4. Analytical Standards
3.1.5. Preparation of the Test Solutions
- (a)
- Dilution of 1.1 mL solution, which contains roughly 100 mg of the dry root P. sidoides extract, in a 10 mL-volumetric flask. The dilution was carried out using methanol and the solution was subjected to sonication and filtration to obtain a 10 mg/mL solution.
- (b)
- Liquid-liquid extraction (LLE) of a 20 mL tincture sample containing roughly 2000 mg of dry root P. sidoides extract with 3 × 20 mL of ethyl acetate. A rotary evaporator was then used to evaporate the combined ethyl acetate extracts to dryness at 45 °C. Finally, the resulting residue weighing 190.9 mg was dissolved in methanol and the volume was adjusted in a 5 mL-volumetric flask.
- (a)
- The first portion (2.015 g) was extracted using a 10 mL methanol solution, sonicated for 5 min, and filtered. Finally, the volume of the solution is adjusted using a 10 mL-volumetric flask to obtain a 10 mg/mL concentration.
- (b)
- The second portion (2.015 g) was suspended in 10 mL of water, sonicated, and extracted using 3 × 20 mL of ethyl acetate. Then, a rotary evaporator was used to evaporate the combined extracts to dryness at 45 °C. The produced residue was reconstituted in methanol and the volume was adjusted in a 5 mL-volumetric flask.
3.1.6. Method Development and Validation
Preparation of the Standard Solution and the Quality Control Samples
Specificity
Linearity
Accuracy and Precision
Robustness
3.2. Assay Procedure for the Determination of the Four Biomolecules in Kalobin, Umca Solutions, and Umca Tablets
3.3. Evaluation of the In Vitro Inhibitory Activity against SARS-CoV-2
3.3.1. The Used Cells and the Viral Strain
3.3.2. MTT Cytotoxicity Assay against Normal Cell Line (Vero E6)
3.3.3. Cytopathic Effect (CPE) Inhibition Assay
3.3.4. Statistical Analysis
3.4. In Silico Studies
3.4.1. Evaluation of Molecular and Pharmacokinetic Properties
3.4.2. Docking Studies
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kolodziej, H. Traditionally used Pelargonium species: Chemistry and biological activity of umckaloabo extracts and their constituents. Curr. Top. Phytochem. 2000, 3, 77–93. [Google Scholar]
- Kayser, O.; Kolodziej, H. Antibacterial activity of extracts and constituents of Pelargonium sidoides and Pelargonium reniforme. Planta Med. 1997, 63, 508–510. [Google Scholar] [CrossRef]
- Careddu, D.; Pettenazzo, A. Pelargonium sidoides extract EPs 7630: A review of its clinical efficacy and safety for treating acute respiratory tract infections in children. Int. J. Gen. Med. 2018, 11, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, A.; Rehman, M.; Alkharfy, K. An alternative approach to minimize the risk of coronavirus (COVID-19) and similar infections. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 4030–4034. [Google Scholar] [PubMed]
- Colling, J.; Groenewald, J.H.; Makunga, N.P. Genetic alterations for increased coumarin production lead to metabolic changes in the medicinally important Pelargonium sidoides DC (Geraniaceae). Metab. Eng. 2010, 12, 561–572. [Google Scholar] [CrossRef]
- Latté, K.P.; Kayser, O.; Tan, N.; Kaloga, M.; Kolodziej, H. Unusual coumarin patterns of Pelargonium Species forming the origin of the traditional herbal medicine Umckaloabo. Z. Nat. C 2000, 55, 528–533. [Google Scholar] [CrossRef]
- Panara, A.; Aalizadeh, R.; Thomaidis, N.S. Chemical characterisation of Pelargonium sidoides root based on LC-QToF-MS non-target screening strategies. Phytochem. Anal. 2022, 33, 40–56. [Google Scholar] [CrossRef]
- Kolodziej, H.; Kiderlen, A.F. In vitro evaluation of antibacterial and immunomodulatory activities of Pelargonium reniforme, Pelargonium sidoides and the related herbal drug preparation EPs 7630. Phytomedicine 2007, 14 (Suppl. 6), 18–26. [Google Scholar] [CrossRef]
- Luypaert, J.; Zhang, M.; Massart, D.L. Feasibility study for the use of near infrared spectroscopy in the qualitative and quantitative analysis of green tea, Camellia Sinensis (L.). Anal. Chim. Acta 2003, 478, 303–312. [Google Scholar] [CrossRef]
- HMPC-Committee-on-Herbal-Medicinal-Products. Assessment Report on Pelargonium sidoides DC and/or Pelargonium reniforme Curt., radix; EMA/HMPC/444251/2015; European Medicines Agency: London, UK, 2018.
- Maree, J.E.; Viljoen, A.M. Phytochemical distinction between Pelargonium sidoides and Pelargonium reniforme—A quality control perspective. S. Afr. J. Bot. 2012, 82, 83–91. [Google Scholar] [CrossRef]
- Kolodziej, H. Fascinating metabolic pools of Pelargonium sidoides and Pelargonium reniforme, traditional and phytomedicinal sources of the herbal medicine Umckaloabo. Phytomedicine 2007, 14 (Suppl. 6), 9–17. [Google Scholar] [CrossRef] [PubMed]
- European-Pharmacopoeia. European Pharmacopoeia, 9th ed.; The Council of Europe: Strasbourg, France, 2016. [Google Scholar]
- Canas, S.; Trindade, C.; Sun, B.; Naves, P. Phenolic compounds involved in pine wilt disease: HPLC-based method development and validation for their quantification. J. Plant Biochem. Biotechnol. 2020, 30, 343–353. [Google Scholar] [CrossRef]
- Dhanani, T.; Shah, S.; Kumar, S. A validated high-performance liquid chromatography method for determination of tannin-related marker constituents gallic acid, corilagin, chebulagic acid, ellagic acid and chebulinic Acid in four Terminalia species from India. J. Chromatogr. Sci. 2015, 53, 625–632. [Google Scholar] [CrossRef]
- Fiori, J.; Pasquini, B.; Caprini, C.; Orlandini, S.; Furlanetto, S.; Gotti, R. Chiral analysis of theanine and catechin in characterization of green tea by cyclodextrin-modified micellar electrokinetic chromatography and high performance liquid chromatography. J. Chromatogr. A 2018, 1562, 115–122. [Google Scholar] [CrossRef]
- Svoboda, P.; Vlčková, H.; Nováková, L. Development and validation of UHPLC–MS/MS method for determination of eight naturally occurring catechin derivatives in various tea samples and the role of matrix effects. J. Pharm. Biomed. Anal. 2015, 114, 62–70. [Google Scholar] [CrossRef]
- Choi, S.-I.; Kwon, H.-Y.; La, I.-J.; Jo, Y.-H.; Han, X.; Men, X.; Lee, S.-J.; Kim, Y.-D.; Seong, G.-S.; Lee, O.-H. Development and validation of an analytical method for deacetylasperulosidic acid, asperulosidic acid, scopolin, asperuloside and scopoletin in fermented Morinda citrifolia L. (Noni). Separations 2021, 8, 80. [Google Scholar] [CrossRef]
- Wang, L.; Wu, H.L.; Yin, X.L.; Hu, Y.; Gu, H.W.; Yu, R.Q. Simultaneous determination of umbelliferone and scopoletin in Tibetan medicine Saussurea laniceps and traditional Chinese medicine Radix Angelicae pubescentis using excitation-emission matrix fluorescence coupled with second-order calibration method. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2017, 170, 104–110. [Google Scholar] [CrossRef]
- Franco, L.; de Oliveira, B.H. Determination of umckalin in commercial tincture and phytopreparations containing Pelargonium sidoides by HPLC: Comparison of sample preparation procedures. Talanta 2010, 81, 1368–1372. [Google Scholar] [CrossRef] [PubMed]
- Masondo, N.A.; Makunga, N.P. Advancement of analytical techniques in some South African commercialized medicinal plants: Current and future perspectives. S. Afr. J. Bot. 2019, 126, 40–57. [Google Scholar] [CrossRef]
- Dömling, A.; Gao, L. Chemistry and Biology of SARS-CoV-2. Chem 2020, 6, 1283–1295. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Lin, D.; Sun, X.; Curth, U.; Drosten, C.; Sauerhering, L.; Becker, S.; Rox, K.; Hilgenfeld, R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science 2020, 368, 409–412. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Zhou, Q.; Li, Y.; Garner, L.V.; Watkins, S.P.; Carter, L.J.; Smoot, J.; Gregg, A.C.; Daniels, A.D.; Jervey, S. Research and development on therapeutic agents and vaccines for COVID-19 and related human coronavirus diseases. ACS Cent. Sci. 2020, 6, 315–331. [Google Scholar] [CrossRef]
- Jia, Z.; Yan, L.; Ren, Z.; Wu, L.; Wang, J.; Guo, J.; Zheng, L.; Ming, Z.; Zhang, L.; Lou, Z. Delicate structural coordination of the Severe Acute Respiratory Syndrome coronavirus Nsp13 upon ATP hydrolysis. Nucleic Acids Res. 2019, 47, 6538–6550. [Google Scholar] [CrossRef]
- Liu, W.; Morse, J.S.; Lalonde, T.; Xu, S. Learning from the past: Possible urgent prevention and treatment options for severe acute respiratory infections caused by 2019-nCoV. Chembiochem 2020, 21, 730–738. [Google Scholar]
- Lung, J.; Lin, Y.S.; Yang, Y.H.; Chou, Y.L.; Shu, L.H.; Cheng, Y.C.; Liu, H.T.; Wu, C.Y. The potential chemical structure of anti-SARS-CoV-2 RNA-dependent RNA polymerase. J. Med. Virol. 2020, 92, 693–697. [Google Scholar] [CrossRef]
- Wu, C.; Liu, Y.; Yang, Y.; Zhang, P.; Zhong, W.; Wang, Y.; Wang, Q.; Xu, Y.; Li, M.; Li, X. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm. Sin. B 2020, 10, 766–788. [Google Scholar] [CrossRef]
- Gurung, A.B.; Ali, M.A.; Lee, J.; Farah, M.A.; Al-Anazi, K.M. Identification of potential SARS-CoV-2 entry inhibitors by targeting the interface region between the spike RBD and human ACE2. J. Infect. Public Health 2021, 14, 227–237. [Google Scholar] [CrossRef]
- NCBI. PubChem Compound Summary for CID 5316862, Umckalin; National Center for Biotechnology Information: Bethesda, MD, USA, 2022.
- NCBI. PubChem Compound Summary for CID 370, Gallic Acid; National Center for Biotechnology Information: Bethesda, MD, USA, 2022.
- NCBI. PubChem Compound Summary for CID 9064, Cianidanol; National Center for Biotechnology Information: Bethesda, MD, USA, 2022.
- NCBI. PubChem Compound Summary for CID 5280460, Scopoletin; National Center for Biotechnology Information: Bethesda, MD, USA, 2022.
- ICH-guideline. Validation of Analytical Procedures: Text and Methodology; International Council for Harmonisation: Geneva, Switzerland, 1995. [Google Scholar]
- Shabir, G.A. Validation of high-performance liquid chromatography methods for pharmaceutical analysis. Understanding the differences and similarities between validation requirements of the US Food and Drug Administration, the US Pharmacopeia and the International Conference on Harmonization. J. Chromatogr. A 2003, 987, 57–66. [Google Scholar] [CrossRef]
- Miller, J.N. Basic statistical methods for analytical chemistry Part 2: Calibration and regression methods. A review. Analyst 1991, 116, 3–14. [Google Scholar] [CrossRef]
- Roth, M.; Fang, L.; Stolz, D.; Tamm, M. Pelargonium sidoides radix extract EPs 7630 reduces rhinovirus infection through modulation of viral binding proteins on human bronchial epithelial cells. PLoS ONE 2019, 14, e0210702. [Google Scholar] [CrossRef]
- Seliem, I.A.; Panda, S.S.; Girgis, A.S.; Moatasim, Y.; Kandeil, A.; Mostafa, A.; Ali, M.A.; Nossier, E.S.; Rasslan, F.; Srour, A.M. New quinoline-triazole conjugates: Synthesis, and antiviral properties against SARS-CoV-2. Bioorg. Chem. 2021, 114, 105117. [Google Scholar] [CrossRef] [PubMed]
- Ikanovic, T.; Sehercehajic, E.; Saric, B.; Tomic, N.; Hadziselimovic, R. In silico analysis of scopoletin interaction with potential SARS-CoV-2 target. In New Technologies, Development and Application IV; Karabegović, I., Ed.; Springer: Berlin/Heidelberg, Germany, 2021; Volume 233. [Google Scholar]
- White, A.G.; Davies-Coleman, M.T.; Ripley, B.S. Measuring and optimising umckalin concentration in wild-harvested and cultivated Pelargonium sidoides (Geraniaceae). S. Afr. J. Bot. 2008, 74, 260–267. [Google Scholar] [CrossRef] [Green Version]
- Trun, W.; Kiderlen, A.F.; Kolodziej, H. Nitric oxide synthase and cytokines gene expression analyses in Leishmania-infected RAW 264.7 cells treated with an extract of Pelargonium sidoides (Eps 7630). Phytomedicine 2006, 13, 570–575. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Zoete, V. A boiled-egg to predict gastrointestinal absorption and brain penetration of small molecules. ChemMedChem 2016, 11, 1117. [Google Scholar] [CrossRef] [PubMed]
- Sliwoski, G.; Kothiwale, S.; Meiler, J.; Lowe, E.W. Computational methods in drug discovery. Pharmacol. Rev. 2014, 66, 334–395. [Google Scholar] [CrossRef] [PubMed]
- Ton, A.T.; Gentile, F.; Hsing, M.; Ban, F.; Cherkasov, A. Rapid identification of potential inhibitors of SARS-CoV-2 main protease by deep docking of 1.3 billion compounds. Mol. Inform. 2020, 39, 2000028. [Google Scholar] [CrossRef]
- Gurung, A.B. In silico structure modelling of SARS-CoV-2 Nsp13 helicase and Nsp14 and repurposing of FDA approved antiviral drugs as dual inhibitors. Gene Rep. 2020, 21, 100860. [Google Scholar]
- Kandeil, A.; Mostafa, A.; Hegazy, R.R.; El-Shesheny, R.; El Taweel, A.; Gomaa, M.R.; Shehata, M.; Elbaset, M.A.; Kayed, A.E.; Mahmoud, S.H.; et al. Immunogenicity and Safety of an Inactivated SARS-CoV-2 Vaccine: Preclinical Studies. Vaccines 2021, 9, 214. [Google Scholar] [CrossRef]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
- Mostafa, A.; Kandeil, A.; Elshaier, Y.A.M.M.; Kutkat, O.; Moatasim, Y.; Rashad, A.A.; Shehata, M.; Gomaa, M.R.; Mahrous, N.; Mahmoud, S.H.; et al. FDA-Approved Drugs with Potent In Vitro Antiviral Activity against Severe Acute Respiratory Syndrome Coronavirus 2. Pharmaceuticals 2020, 13, 443. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed] [Green Version]





| Parameter * | Gallic Acid | Catechin | Scopoletin | Umckalin |
|---|---|---|---|---|
| Concentration range (µg/mL) | 0.2–1 | 0.2–1 | 0.1–1 | 0.1–1 |
| Correlation coefficient () | 0.9995 | 0.9999 | 0.9998 | 0.9995 |
| Slope | 129,746.6 | 164,166.5 | 162,561.9 | 188,550.2 |
| Intercept | 1655.6 | 2039.8 | 2223.6 | 3046.9 |
| , S.D. of the residuals | 1059.3 | 670.3 | 827.0 | 1456.5 |
| , S.D. of the intercept | 943.78 | 597.28 | 589.57 | 1038.3 |
| , S.D. of the slope | 1700.6 | 1076.2 | 1159.9 | 2042.8 |
| S.D. | 1.95 | 1.08 | 1.87 | 2.12 |
| %RSD a | 1.95 | 1.08 | 1.86 | 2.11 |
| %Error b | 0.873 | 0.484 | 0.762 | 0.866 |
| LOD c (µg/mL) | 0.024 | 0.012 | 0.012 | 0.018 |
| LOQ d (µg/mL) | 0.073 | 0.036 | 0.036 | 0.055 |
| Responses | Concentrations (μg/mL) * | |||
|---|---|---|---|---|
| Gallic Acid | Catechin | Scopoletin | Umckalin | |
| Response at 0.2 µg/mL | Concentration:0.2 µg/mL | |||
| 28,551 | 34,214 | 34,084 | 40,077 | |
| 28,740 | 34,350 | 34,079 | 40,223 | |
| 28,340 | 34,153 | 34,090 | 40,122 | |
| X | 28,543.67 | 34,239.00 | 34,084.33 | 40,140.67 |
| SD | 200.10 | 100.85 | 5.51 | 74.77 |
| %RSD | 0.70 | 0.29 | 0.02 | 0.19 |
| Conc. Found *(µg/mL) | 0.204 | 0.196 | 0.196 | 0.196 |
| % Found | 102.10 | 98.20 | 98.00 | 98.40 |
| Response at 0.3 µg/mL | Concentration: 0.3 µg/mL | |||
| 40,552 | 51,184 | 51,610 | 60,560 | |
| 41,019 | 51,435 | 51,633 | 60,443 | |
| 40,391 | 51,099 | 51,621 | 60,492 | |
| X | 40,654.00 | 51,239.33 | 51,621.33 | 60,498.33 |
| SD | 326.19 | 174.70 | 11.50 | 58.76 |
| %RSD | 0.80 | 0.34 | 0.02 | 0.10 |
| Conc. Found *(µg/mL) | 0.307 | 0.298 | 0.304 | 0.304 |
| % Found | 102.33 | 99.53 | 101.30 | 101.57 |
| Response at 0.5 µg/mL | Concentration: 0.5 µg/mL | |||
| 65,527 | 75,560 | 72,072 | 83,973 | |
| 65,256 | 75,118 | 72,253 | 84,031 | |
| 64,322 | 75,233 | 72,165 | 83,992 | |
| X | 65,035.00 | 75,303.67 | 72,163.33 | 83,998.67 |
| SD | 632.17 | 229.32 | 90.51 | 29.57 |
| %RSD | 0.97 | 0.30 | 0.13 | 0.04 |
| Conc. Found *(µg/mL) | 0.491 | 0.505 | 0.504 | 0.507 |
| % Found | 98.24 | 101.00 | 100.80 | 101.56 |
| ± SD | 100.89 ± 2.2 | 99.57 ± 1.4 | 100.03 ± 1.7 | 100.51 ± 1.8 |
| Test Sample | CC50 (μM) a | IC50 (μM) b | Selectivity Index (SI) | Concentration of Investigated Biomolecules (μg/mL) ** | |||||
|---|---|---|---|---|---|---|---|---|---|
| Kalobin®-S | Umca®-S | Umca®-T | |||||||
| Ethyl Acetate | Methanol | Ethyl Acetate | Methanol | Ethyl Acetate | Methanol | ||||
| Gallic acid | 439.7 ± 0.065 | 96.41 ± 0.030 | 4.6 | 89.54 | 22.00 | 188.71 | 98.91 | 146.66 | 23.72 |
| Catechin | 52.76 ± 0.079 | 58.55 ± 0.088 | 0.9 | 14.05 | 10.96 | 39.09 | 0 | 11.26 | 0 |
| Scopoletin | 253.1 ± 0.45 | 17.79 ± 0.91 | 14.1 | 14.86 | 0 | 6.31 | 0 | 27.19 | 0 |
| Umckalin | 164.1 ± 0.54 | 311.6 ± 0.043 | 0.5 | 121.07 | 3.09 | 79.46 | 2.86 | 126.33 | 4.54 |
| P. sidoides root extract | 87.25 ± 0.093 c | 13.79 ± 0.034 c | 6.3 | ||||||
| Chloroquine * | 377.7 | 22.7 | 16.64 | ||||||
| Hydroxychloroquine * | 356 | 32.8 | 10.85 | ||||||
| Compound | Binding Energy (Kcal/mol) | ||||
|---|---|---|---|---|---|
| Mpro | PLpro | Nsp13 | RdRp | The Interface of RBD of Spike Protein with Its Human ACE2 Receptor | |
| Catechin | −7.2 | −6.1 | −7.2 | −6.4 | −7.7 |
| Gallic acid | −5.1 | −4.5 | −5.7 | −5.4 | −6.4 |
| Umckalin | −5.3 | −4.8 | −6.6 | −5.4 | −6.3 |
| Scopoletin | −5.2 | −4.9 | −6.5 | −5.8 | −6.4 |
| Reference inhibitor | −4.9 | −6.7 | −5.7 | −8.9 | −6.6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alossaimi, M.A.; Alzeer, M.A.; Abdel Bar, F.M.; ElNaggar, M.H. Pelargonium sidoides Root Extract: Simultaneous HPLC Separation, Determination, and Validation of Selected Biomolecules and Evaluation of SARS-CoV-2 Inhibitory Activity. Pharmaceuticals 2022, 15, 1184. https://0-doi-org.brum.beds.ac.uk/10.3390/ph15101184
Alossaimi MA, Alzeer MA, Abdel Bar FM, ElNaggar MH. Pelargonium sidoides Root Extract: Simultaneous HPLC Separation, Determination, and Validation of Selected Biomolecules and Evaluation of SARS-CoV-2 Inhibitory Activity. Pharmaceuticals. 2022; 15(10):1184. https://0-doi-org.brum.beds.ac.uk/10.3390/ph15101184
Chicago/Turabian StyleAlossaimi, Manal A., May A. Alzeer, Fatma M. Abdel Bar, and Mai H. ElNaggar. 2022. "Pelargonium sidoides Root Extract: Simultaneous HPLC Separation, Determination, and Validation of Selected Biomolecules and Evaluation of SARS-CoV-2 Inhibitory Activity" Pharmaceuticals 15, no. 10: 1184. https://0-doi-org.brum.beds.ac.uk/10.3390/ph15101184