CRISPR/dCas9-Based Systems: Mechanisms and Applications in Plant Sciences
Abstract
:1. Introduction
2. The CRISPR/Cas System: Discovery and Principle
3. The Current CRISPR/Cas9 System and Strategies to Mitigate Off-Target Effects
- (a)
- bioinformatics selection and modification of sgRNA;
- (b)
- finetuning expression of CRISPR components;
- (c)
- use of Cas9 variants and orthologs;
- (d)
- utilization of heterologous nucleases in the CRISPR system;
- (e)
- alternative CRISPR approaches.
3.1. Bioinformatics Selection and Modification of sgRNA
3.2. Finetuning Expression of CRISPR Components
3.3. Use of Cas9 Variants and Orthologues
3.4. Utilization of Heterologous Nucleases
3.5. Alternative CRISPR Approaches
4. Inactive CRISPR-Associated Nucleases: A Transcriptional Regulator
4.1. sgRNA
4.2. dCas9
4.3. Transcriptional Effectors
5. Strategies for Programable Transcriptional Regulations in Plants
5.1. Multiple sgRNAs
5.2. Modification of CRISPR/dCas9 Components
5.3. Transcriptional Regulation Toolbox
5.4. Plant Specific Transcriptional Effectors
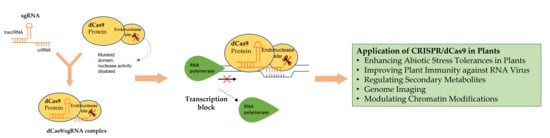
6. Application of CRISPR/dCas9 in Plants
6.1. Enhancing Abiotic Stress Tolerances in Plants
6.2. Improving Plant Immunity against RNA Virus
6.3. Regulation of Secondary Metabolites
6.4. Other Applications of CRISPR/dCas9
7. Challenges and Issues of CRISPR/dCas9
8. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Guigo, R.; De Hoon, M. Recent advances in functional genome analysis. F1000Research 2018, 7, 1968. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yadav, B.S.; Tripathi, V. Recent Advances in the System Biology-based Target Identification and Drug Discovery. Curr. Top. Med. Chem. 2018, 18, 1737–1744. [Google Scholar] [CrossRef]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Fire, A.; Xu, S.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998, 391, 806–811. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Gupta, M.; Pandher, S.; Kaur, G.; Goel, N.; Rathore, P. Author Correction: Using de novo transcriptome assembly and analysis to study RNAi in Phenacoccus solenopsis Tinsley (Hemiptera: Pseudococcidae). Sci. Rep. 2020, 10, 1. [Google Scholar] [CrossRef] [Green Version]
- Hanly, A.; Karagiannis, J.; Lu, Q.S.M.; Tian, L.; Hannoufa, A. Characterization of the Role of SPL9 in Drought Stress Tolerance in Medicago sativa. Int. J. Mol. Sci. 2020, 21, 6003. [Google Scholar] [CrossRef]
- Mamta, B.; Rajam, M.V. RNAi technology: A new platform for crop pest control. Physiol. Mol. Biol. Plants 2017, 23, 487–501. [Google Scholar] [CrossRef]
- Boettcher, M.; McManus, M. Choosing the right tool for the job: RNAi, TALEN, or CRISPR. Mol. Cell 2015, 58, 575–585. [Google Scholar] [CrossRef] [Green Version]
- Wada, N.; Ueta, R.; Osakabe, Y.; Osakabe, K. Precision genome editing in plants: State-of-the-art in CRISPR/Cas9-based genome engineering. BMC Plant Biol. 2020, 20, 1–12. [Google Scholar] [CrossRef]
- Xu, X.; Qi, L.S. A CRISPR–dCas toolbox for genetic engineering and synthetic biology. J. Mol. Biol. 2019, 431, 34–47. [Google Scholar] [CrossRef]
- Ishino, Y.; Shinagawa, H.; Makino, K.; Amemura, M.; Nakata, A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J. Bacteriol. 1987, 169, 5429–5433. [Google Scholar] [CrossRef] [Green Version]
- Mojica, F.J.; Ferrer, C.; Juez, G.; Rodriguez-Valera, F. Long stretches of short tandem repeats are present in the largest replicons of the Archaea Haloferax mediterranei and Haloferax volcanii and could be involved in replicon partitioning. Mol. Microbiol. 1995, 17, 85–93. [Google Scholar] [CrossRef]
- Mojica, F.J.M.; Diez-Villasenor, C.; Soria, E.; Juez, G. Biological significance of a family of regularly spaced repeats in the genomes of Archaea, Bacteria and mitochondria. Mol. Microbiol. 2000, 36, 244–246. [Google Scholar] [CrossRef]
- Jansen, R.; Van Embden, J.D.A.; Gaastra, W.; Schouls, L.M. Identification of genes that are associated with DNA repeats in prokaryotes. Mol. Microbiol. 2002, 43, 1565–1575. [Google Scholar] [CrossRef]
- Bolotin, A.; Quinquis, B.; Sorokine, A.; Ehrlich, S.D. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology 2005, 151, 2551–2561. [Google Scholar] [CrossRef] [Green Version]
- Pourcel, C.; Salvignol, G.; Vergnaud, G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology 2005, 151, 653–663. [Google Scholar] [CrossRef] [Green Version]
- Mojica, F.J.M.; Soria, E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol. 2005, 60, 174–182. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Grishin, N.V.; Shabalina, S.A.; Wolf, Y.I.; Koonin, E.V. A putative RNA-interference-based immune system in prokaryotes: Computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol. Direct 2006, 1, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barrangou, R.; Fremaux, C.; Deveau, H.; Richards, M.; Boyaval, P.; Moineau, S.; Romero, D.A.; Horvath, P. CRISPR provides acquired resistance against viruses in prokaryotes. Science 2007, 315, 1709–1712. [Google Scholar] [CrossRef] [PubMed]
- Deveau, H.; Barrangou, R.; Garneau, J.E.; Labonte, J.; Fremaux, C.; Boyaval, P.; Romero, D.A.; Horvath, P.; Moineau, S. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J. Bacteriol. 2008, 190, 1390–1400. [Google Scholar] [CrossRef] [Green Version]
- Horvath, P.; Romero, D.A.; Coûteé-Monvoisin, A.-C.; Richards, M.; Deveau, H.; Moineau, S.; Boyaval, P.; Fremaux, C.; Barrangou, R. Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus. J. Bacteriol. 2008, 190, 1401–1412. [Google Scholar] [CrossRef] [Green Version]
- He, L.; James, M.S.J.; Radovcic, M.; Ivancic-Bace, I.; Bolt, E.L. Cas3 Protein—A review of a multi-tasking machine. Genes 2020, 11, 208. [Google Scholar] [CrossRef] [Green Version]
- Dorsey, B.W.; Huang, L.; Mondragón, A. Structural organization of a Type III-A CRISPR effector subcomplex determined by X-ray crystallography and cryo-EM. Nucleic Acids Res. 2019, 47, 3765–3783. [Google Scholar] [CrossRef] [Green Version]
- Deltcheva, E.; Chylinski, K.; Sharma, C.M.; Gonzales, K.; Chao, Y.; Pirzada, Z.A.; Eckert, M.R.; Vogel, J.; Charpentier, E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 2011, 471, 602–607. [Google Scholar] [CrossRef] [Green Version]
- Gleditzsch, D.; Pausch, P.; Esparza, H.M.; Özcan, A.; Guo, X.; Bange, G.; Randau, L. PAM identification by CRISPR-Cas effector complexes: Diversified mechanisms and structures. RNA Biol. 2018, 16, 504–517. [Google Scholar] [CrossRef] [PubMed]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided dna endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef] [PubMed]
- De Pater, S.; Klemann, B.J.P.M.; Hooykaas, P.J.J. True gene-targeting events by CRISPR/Cas-induced DSB repair of the PPO locus with an ectopically integrated repair template. Sci. Rep. 2018, 8, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Zaboikin, M.; Zaboikina, T.; Freter, C.; Srinivasakumar, N. Non-homologous end joining and homology directed DNA repair frequency of double-stranded breaks introduced by Genome editing reagents. PLoS ONE 2017, 12, e0169931. [Google Scholar] [CrossRef] [PubMed]
- Ricroch, A.; Clairand, P.; Harwood, W. Use of CRISPR systems in plant genome editing: Toward new opportunities in agriculture. Emerg. Top. Life Sci. 2017, 1, 169–182. [Google Scholar] [CrossRef] [Green Version]
- Leblanc, C.; Zhang, F.; Mendez, J.; Lozano, Y.; Chatpar, K.; Irish, V.F.; Jacob, Y. Increased efficiency of targeted mutagenesis by CRISPR/Cas9 in plants using heat stress. Plant J. 2017, 93, 377–386. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Alptekin, B.; Budak, H. CRISPR/Cas9 genome editing in wheat. Funct. Integr. Genom. 2017, 18, 31–41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, J.; Gao, H.; Wang, H.; Lafitte, H.R.; Archibald, R.L.; Yang, M.; Hakimi, S.M.; Mo, H.; Habben, J.E. ARGOS8 variants generated by CRISPR-Cas9 improve maize grain yield under field drought stress conditions. Plant Biotechnol. J. 2016, 15, 207–216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, W.; Pan, Q.; He, F.; Akhunova, A.; Chao, S.; Trick, H.; Akhunov, E. Transgenerational CRISPR-Cas9 activity facilitates multiplex gene editing in allopolyploid wheat. CRISPR J. 2018, 1, 65–74. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Peng, Z.; Long, J.; Sosso, D.; Liu, B.; Eom, J.-S.; Huang, S.; Liu, S.; Cruz, C.V.; Frommer, W.; et al. Gene targeting by the TAL effector PthXo2 reveals cryptic resistance gene for bacterial blight of rice. Plant J. 2015, 82, 632–643. [Google Scholar] [CrossRef]
- Zhang, Z.; Ge, X.; Luo, X.; Wang, P.; Fan, Q.; Hu, G.; Xiao, J.; Li, F.; Wu, J. Simultaneous editing of two copies of Gh14-3-3d confers enhanced transgene-clean plant defense against verticillium dahliae in allotetraploid upland cotton. Front. Plant Sci. 2018, 9, 842. [Google Scholar] [CrossRef]
- Chandrasekaran, J.; Brumin, M.; Wolf, D.; Leibman, D.; Klap, C.; Pearlsman, M.; Sherman, A.; Arazi, T.; Gal-On, A. Development of broad virus resistance in non-transgenic cucumber using CRISPR/Cas9 technology. Mol. Plant Pathol. 2016, 17, 1140–1153. [Google Scholar] [CrossRef] [Green Version]
- Oliva, R.; Ji, C.; Atienza-Grande, G.; Huguet-Tapia, J.C.; Perez-Quintero, A.; Li, T.; Eom, J.-S.; Li, C.; Nguyen, H.; Liu, B.; et al. Broad-spectrum resistance to bacterial blight in rice using genome editing. Nat. Biotechnol. 2019, 37, 1344–1350. [Google Scholar] [CrossRef] [Green Version]
- Zheng, S.; Ye, C.; Lu, J.; Liufu, J.; Lin, L.; Dong, Z.; Li, J.; Zhuang, C. Improving the rice photosynthetic efficiency and yield by editing OsHXK1 via CRISPR/Cas9 system. Int. J. Mol. Sci. 2021, 22, 9554. [Google Scholar] [CrossRef]
- Scheben, A.; Wolter, F.; Batley, J.; Puchta, H.; Edwards, D. Towards CRISPR/Cas crops—Bringing together genomics and genome editing. New Phytol. 2017, 216, 682–698. [Google Scholar] [CrossRef] [Green Version]
- Kaur, N.; Alok, A.; Shivani, M.; Kumar, P.; Kaur, N.; Awasthi, P.; Chaturvedi, S.; Pandey, P.; Pandey, A.; Pandey, A.K.; et al. CRISPR/Cas9 directed editing of lycopene epsilon-cyclase modulates metabolic flux for β-carotene biosynthesis in banana fruit. Metab. Eng. 2020, 59, 76–86. [Google Scholar] [CrossRef] [PubMed]
- Dong, O.X.; Yu, S.; Jain, R.; Zhang, N.; Duong, P.Q.; Butler, C.; Li, Y.; Lipzen, A.; Martin, J.A.; Barry, K.; et al. Marker-free carotenoid-enriched rice generated through targeted gene insertion using CRISPR-Cas9. Nat. Commun. 2020, 11, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Ahmad, S.; Tang, L.; Shahzad, R.; Mawia, A.M.; Rao, G.S.; Jamil, S.; Wei, C.; Sheng, Z.; Shao, G.; Wei, X.; et al. CRISPR-based crop improvements: A way forward to achieve zero hunger. J. Agric. Food Chem. 2021, 69, 8307–8323. [Google Scholar] [CrossRef]
- Kanchiswamy, C.N.; Maffei, M.; Malnoy, M.; Velasco, R.; Kim, J.-S. Fine-tuning next-generation genome editing tools. Trends Biotechnol. 2016, 34, 562–574. [Google Scholar] [CrossRef]
- Troadec, M.-B.; Pages, J.-C. Where are we with unintended effects in genome editing applications from DNA to phenotype: Focus on plant applications. Transgenic Res. 2019, 28, 125–133. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Cheng, X.; Shan, Q.; Zhang, Y.; Liu, J.; Gao, C.; Qiu, J.-L. Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 2014, 32, 947–951. [Google Scholar] [CrossRef]
- Ren, X.; Yang, Z.; Xu, J.; Sun, J.; Mao, D.; Hu, Y.; Yang, S.-J.; Qiao, H.-H.; Wang, X.; Hu, Q.; et al. Enhanced specificity and efficiency of the CRISPR/Cas9 system with optimized sgRNA parameters in drosophila. Cell Rep. 2014, 9, 1151–1162. [Google Scholar] [CrossRef] [Green Version]
- Ran, F.A.; Hsu, P.; Wright, J.D.; Agarwala, V.; Scott, D.A.; Zhang, F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013, 8, 2281–2308. [Google Scholar] [CrossRef] [Green Version]
- Rose, J.C.; Popp, N.A.; Richardson, C.D.; Stephany, J.J.; Mathieu, J.; Wei, C.T.; Corn, J.E.; Maly, D.J.; Fowler, D.M. Suppression of unwanted CRISPR-Cas9 editing by co-administration of catalytically inactivating truncated guide RNAs. Nat. Commun. 2020, 11, 1–11. [Google Scholar] [CrossRef]
- Ryan, M.; Taussig, D.; Steinfeld, I.; Phadnis, S.M.; Lunstad, B.D.; Singh, M.; Vuong, X.; Okochi, K.D.; McCaffrey, R.; Olesiak, M.; et al. Improving CRISPR–Cas specificity with chemical modifications in single-guide RNAs. Nucleic Acids Res. 2017, 46, 792–803. [Google Scholar] [CrossRef]
- Yin, H.; Song, C.-Q.; Suresh, S.; Kwan, S.-Y.; Wu, Q.; Walsh, S.; Ding, J.; Bogorad, R.L.; Zhu, L.J.; Wolfe, S.A.; et al. Partial DNA-guided Cas9 enables genome editing with reduced off-target activity. Nat. Chem. Biol. 2018, 14, 311–316. [Google Scholar] [CrossRef] [Green Version]
- Hendel, A.; Bak, R.; Clark, J.T.; Kennedy, A.B.; Ryan, D.E.; Roy, S.; Steinfeld, I.; Lunstad, B.D.; Kaiser, R.J.; Wilkens, A.B.; et al. Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nat. Biotechnol. 2015, 33, 985–989. [Google Scholar] [CrossRef]
- Yin, H.; Song, C.-Q.; Suresh, S.; Wu, Q.; Walsh, S.; Rhym, L.H.; Mintzer, E.; Bolukbasi, M.F.; Zhu, L.J.; Kauffman, K.; et al. Structure-guided chemical modification of guide RNA enables potent non-viral in vivo genome editing. Nat. Biotechnol. 2017, 35, 1179–1187. [Google Scholar] [CrossRef] [PubMed]
- Cromwell, C.R.; Sung, K.; Park, J.; Krysler, A.R.; Jovel, J.; Kim, S.K.; Hubbard, B.P. Incorporation of bridged nucleic acids into CRISPR RNAs improves Cas9 endonuclease specificity. Nat. Commun. 2018, 9, 1–11. [Google Scholar] [CrossRef]
- Rahdar, M.; McMahon, M.A.; Prakash, T.P.; Swayze, E.E.; Bennett, C.F.; Cleveland, D.W. Synthetic CRISPR RNA-Cas9–guided genome editing in human cells. Proc. Natl. Acad. Sci. USA 2015, 112, E7110–E7117. [Google Scholar] [CrossRef] [Green Version]
- Hsu, P.; Scott, D.A.; Weinstein, J.; Ran, F.A.; Konermann, S.; Agarwala, V.; Li, Y.; Fine, E.; Wu, X.; Shalem, O.; et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013, 31, 827–832. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishna, S.; Dad, A.-B.K.; Beloor, J.; Gopalappa, R.; Lee, S.-K.; Kim, H. Gene disruption by cell-penetrating peptide-mediated delivery of Cas9 protein and guide RNA. Genome Res. 2014, 24, 1020–1027. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.; Kim, D.; Cho, S.W.; Kim, J. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014, 24, 1012–1019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gasiunas, G.; Young, J.K.; Karvelis, T.; Kazlauskas, D.; Urbaitis, T.; Jasnauskaite, M.; Grusyte, M.M.; Paulraj, S.; Wang, P.-H.; Hou, Z.; et al. A catalogue of biochemically diverse CRISPR-Cas9 orthologs. Nat. Commun. 2020, 11, 1–10. [Google Scholar] [CrossRef]
- Ran, F.A.; Cong, L.; Yan, W.X.; Scott, D.A.; Gootenberg, J.; Kriz, A.J.; Zetsche, B.; Shalem, O.; Wu, X.; Makarova, K.S.; et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature 2015, 520, 186–191. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.; Cradick, T.; Bao, G. The Neisseria meningitidis CRISPR-Cas9 System enables specific genome editing in mammalian cells. Mol. Ther. 2016, 24, 645–654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steinert, J.; Schiml, S.; Fauser, F.; Puchta, H. Highly efficient heritable plant genome engineering using Cas9 orthologues from Streptococcus thermophilus and Staphylococcus aureus. Plant J. 2015, 84, 1295–1305. [Google Scholar] [CrossRef] [PubMed]
- Müller, M.; Lee, C.; Gasiunas, G.; Davis, T.H.; Cradick, T.; Siksnys, V.; Bao, G.; Cathomen, T.; Mussolino, C. Streptococcus thermophilus CRISPR-Cas9 Systems enable specific editing of the human genome. Mol. Ther. 2016, 24, 636–644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nishimasu, H.; Shi, X.; Ishiguro, S.; Gao, L.; Hirano, S.; Okazaki, S.; Noda, T.; Abudayyeh, O.O.; Gootenberg, J.S.; Mori, H.; et al. Engineered CRISPR-Cas9 nuclease with expanded targeting space. Science 2018, 361, 1259–1262. [Google Scholar] [CrossRef] [PubMed]
- Ge, Z.; Zheng, L.; Zhao, Y.; Jiang, J.; Zhang, E.J.; Liu, T.; Gu, H.; Qu, L. Engineered xCas9 and SpCas9-NG variants broaden PAM recognition sites to generate mutations in Arabidopsis plants. Plant Biotechnol. J. 2019, 17, 1865–1867. [Google Scholar] [CrossRef] [Green Version]
- Hua, K.; Tao, X.; Han, P.; Wang, R.; Zhu, J.-K. Genome engineering in rice using Cas9 variants that recognize NG PAM sequences. Mol. Plant 2019, 12, 1003–1014. [Google Scholar] [CrossRef]
- Slaymaker, I.M.; Gao, L.; Zetsche, B.; Scott, D.A.; Yan, W.X.; Zhang, F. Rationally engineered Cas9 nucleases with improved specificity. Science 2015, 351, 84–88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.; Dagdas, Y.S.; Kleinstiver, B.; Welch, M.M.; Sousa, A.A.; Harrington, L.B.; Sternberg, S.H.; Joung, J.K.; Yildiz, A.; Doudna, J.A. Enhanced proofreading governs CRISPR–Cas9 targeting accuracy. Nature 2017, 550, 407–410. [Google Scholar] [CrossRef] [Green Version]
- Kleinstiver, B.; Pattanayak, V.; Prew, M.S.; Tsai, S.Q.; Nguyen, N.T.; Zheng, Z.; Joung, J.K. High-fidelity CRISPR–Cas9 nucleases with no detectable genome-wide off-target effects. Nature 2016, 529, 490–495. [Google Scholar] [CrossRef] [Green Version]
- Zhang, D.; Zhang, H.; Li, T.; Chen, K.; Qiu, J.-L.; Gao, C. Perfectly matched 20-nucleotide guide RNA sequences enable robust genome editing using high-fidelity SpCas9 nucleases. Genome Biol. 2017, 18, 191. [Google Scholar] [CrossRef] [Green Version]
- Dugar, G.; Leenay, R.T.; Eisenbart, S.K.; Bischler, T.; Aul, B.U.; Beisel, C.L.; Sharma, C.M. CRISPR RNA-Dependent Binding and Cleavage of Endogenous RNAs by the Campylobacter jejuni Cas9. Mol. Cell 2018, 69, 893–905.e7. [Google Scholar] [CrossRef] [Green Version]
- Shen, B.; Zhang, W.; Zhang, J.; Zhou, J.; Wang, J.; Chen, L.; Wang, L.; Hodgkins, A.; Iyer, V.; Huang, X.; et al. Efficient genome modification by CRISPR-Cas9 nickase with minimal off-target effects. Nat. Methods 2014, 11, 399–402. [Google Scholar] [CrossRef]
- Schiml, S.; Fauser, F.; Puchta, H. Repair of adjacent single-strand breaks is often accompanied by the formation of tandem sequence duplications in plant genomes. Proc. Natl. Acad. Sci. USA 2016, 113, 7266–7271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cho, S.W.; Kim, S.; Kim, Y.; Kweon, J.; Kim, H.S.; Bae, S.; Kim, J.-S. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res. 2013, 24, 132–141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frock, R.; Hu, J.; Meyers, R.; Ho, Y.-J.; Kii, E.; Alt, F.W. Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases. Nat. Biotechnol. 2014, 33, 179–186. [Google Scholar] [CrossRef] [PubMed]
- Guilinger, J.P.; Thompson, D.B.; Liu, D.R. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat. Biotechnol. 2014, 32, 577–582. [Google Scholar] [CrossRef]
- Mali, P.; Aach, J.; Stranges, P.; Esvelt, K.; Moosburner, M.; Kosuri, S.; Yang, L.; Church, G.M. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 2013, 31, 833–838. [Google Scholar] [CrossRef] [PubMed]
- Tsai, S.Q.; Wyvekens, N.; Khayter, C.; Foden, J.A.; Thapar, V.; Reyon, D.; Goodwin, M.J.; Aryee, M.J.; Joung, J.K. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat. Biotechnol. 2014, 32, 569–576. [Google Scholar] [CrossRef] [Green Version]
- Komor, A.; Kim, Y.B.; Packer, M.S.; Zuris, J.A.; Liu, D.R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 2016, 533, 420–424. [Google Scholar] [CrossRef] [Green Version]
- Marzec, M.; Hensel, G. Targeted base editing systems are available for plants. Trends Plant Sci. 2018, 23, 955–957. [Google Scholar] [CrossRef]
- Molla, K.A.; Yang, Y. CRISPR/Cas-mediated base editing: Technical considerations and practical applications. Trends Biotechnol. 2019, 37, 1121–1142. [Google Scholar] [CrossRef]
- Shimatani, Z.; Kashojiya, S.; Takayama, M.; Terada, R.; Arazoe, T.; Ishii, H.; Teramura, H.; Yamamoto, T.; Komatsu, H.; Miura, K.; et al. Targeted base editing in rice and tomato using a CRISPR-Cas9 cytidine deaminase fusion. Nat. Biotechnol. 2017, 35, 441–443. [Google Scholar] [CrossRef]
- Anzalone, A.V.; Randolph, P.B.; Davis, J.R.; Sousa, A.A.; Koblan, L.; Levy, J.M.; Chen, P.; Wilson, C.; Newby, G.A.; Raguram, A.; et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 2019, 576, 149–157. [Google Scholar] [CrossRef]
- Lin, Q.; Zong, Y.; Xue, C.; Wang, S.; Jin, S.; Zhu, Z.; Wang, Y.; Anzalone, A.V.; Raguram, A.; Doman, J.L.; et al. Prime genome editing in rice and wheat. Nat. Biotechnol. 2020, 38, 582–585. [Google Scholar] [CrossRef]
- Tang, X.; Sretenovic, S.; Ren, Q.; Jia, X.; Li, M.; Fan, T.; Yin, D.; Xiang, S.; Guo, Y.; Liu, L.; et al. Plant prime editors enable precise gene editing in rice cells. Mol. Plant 2020, 13, 667–670. [Google Scholar] [CrossRef]
- Li, H.; Li, J.; Chen, J.; Yan, L.; Xia, L. Precise modifications of both exogenous and endogenous genes in rice by prime editing. Mol. Plant 2020, 13, 671–674. [Google Scholar] [CrossRef]
- Nakamura, M.; Gao, Y.; Dominguez, A.A.; Qi, L.S. CRISPR technologies for precise epigenome editing. Nature 2021, 23, 11–22. [Google Scholar] [CrossRef]
- Galonska, C.; Charlton, J.; Mattei, A.L.; Donaghey, J.; Clement, K.; Gu, H.; Mohammad, A.W.; Stamenova, E.K.; Cacchiarelli, D.; Klages, S.; et al. Genome-wide tracking of dCas9-methyltransferase footprints. Nat. Commun. 2018, 9, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stepper, P.; Kungulovski, G.; Jurkowska, R.Z.; Chandra, T.; Krueger, F.; Reinhardt, R.; Reik, W.; Jeltsch, A.; Jurkowski, T.P. Efficient targeted DNA methylation with chimeric dCas9–Dnmt3a–Dnmt3L methyltransferase. Nucleic Acids Res. 2016, 45, 1703–1713. [Google Scholar] [CrossRef] [PubMed]
- Qi, L.S.; Larson, M.H.; Gilbert, L.; Doudna, J.A.; Weissman, J.S.; Arkin, A.; Lim, W.A. Repurposing CRISPR as an RNA-Guided Platform for Sequence-Specific Control of Gene Expression. Cell 2013, 152, 1173–1183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khatodia, S.; Bhatotia, K.; Passricha, N.; Khurana, S.M.P.; Tuteja, N. The CRISPR/Cas genome-editing tool: Application in improvement of crops. Front. Plant Sci. 2016, 7, 506. [Google Scholar] [CrossRef] [Green Version]
- O’Geen, H.; Ren, C.; Nicolet, C.M.; Perez, A.A.; Halmai, J.; Le, V.M.; Mackay, J.P.; Farnham, P.J.; Segal, D.J. dCas9-based epigenome editing suggests acquisition of histone methylation is not sufficient for target gene repression. Nucleic Acids Res. 2017, 45, 9901–9916. [Google Scholar] [CrossRef] [Green Version]
- Uniyal, A.P.; Mansotra, K.; Yadav, S.K.; Kumar, V. An overview of designing and selection of sgRNAs for precise genome editing by the CRISPR-Cas9 system in plants. 3 Biotech 2019, 9, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, L.; Larson, M.H.; Morsut, L.; Liu, Z.; Brar, G.A.; Torres, S.E.; Stern-Ginossar, N.; Brandman, O.; Whitehead, E.H.; Doudna, J.A.; et al. CRISPR-Mediated Modular RNA-guided regulation of transcription in eukaryotes. Cell 2013, 154, 442–451. [Google Scholar] [CrossRef] [Green Version]
- Li, X.-T.; Jun, Y.; Erickstad, M.J.; Brown, S.D.; Parks, A.; Court, D.L.; Jun, S. tCRISPRi: Tunable and reversible, one-step control of gene expression. Sci. Rep. 2016, 6, 39076. [Google Scholar] [CrossRef] [PubMed]
- Piatek, A.; Ali, Z.; Baazim, H.; Li, L.; Abulfaraj, A.; AlShareef, S.; Aouida, M.; Mahfouz, M.M. RNA-guided transcriptional regulationin plantavia synthetic dCas9-based transcription factors. Plant Biotechnol. J. 2014, 13, 578–589. [Google Scholar] [CrossRef] [PubMed]
- Adli, M. The CRISPR tool kit for genome editing and beyond. Nat. Commun. 2018, 9, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Thakore, P.I.; Black, J.B.; Hilton, I.B.; Gersbach, C.A. Editing the epigenome: Technologies for programmable transcription and epigenetic modulation. Nat. Methods 2016, 13, 127–137. [Google Scholar] [CrossRef]
- Casas-Mollano, J.A.; Zinselmeier, M.H.; Erickson, S.E.; Smanski, M.J. CRISPR-Cas activators for engineering gene expression in higher eukaryotes. CRISPR J. 2020, 3, 350–364. [Google Scholar] [CrossRef]
- Jain, M. Function genomics of abiotic stress tolerance in plants: A CRISPR approach. Front. Plant Sci. 2015, 6, 375. [Google Scholar] [CrossRef] [Green Version]
- Osakabe, Y.; Sugano, S.S.; Osakabe, K. Genome engineering of woody plants: Past, present and future. J. Wood Sci. 2016, 62, 217–225. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Moschou, P.N. Phenotypic novelty by CRISPR in plants. Dev. Biol. 2018, 435, 170–175. [Google Scholar] [CrossRef] [PubMed]
- Conaway, J.W. Introduction to theme “Chromatin, epigenetics, and transcription”. Annu. Rev. Biochem. 2012, 81, 61–64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kribelbauer, J.; Lu, X.-J.; Rohs, R.; Mann, R.S.; Bussemaker, H.J. Toward a mechanistic understanding of DNA methylation readout by transcription factors. J. Mol. Biol. 2019, 432, 1801–1815. [Google Scholar] [CrossRef] [PubMed]
- Omalley, R.; Huang, S.-S.C.; Song, L.; Lewsey, M.; Bartlett, A.; Nery, J.R.; Galli, M.; Gallavotti, A.; Ecker, J.R. Cistrome and epicistrome features shape the regulatory DNA landscape. Cell 2016, 165, 1280–1292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bikard, D.; Jiang, W.; Samai, P.; Hochschild, A.; Zhang, F.; Marraffini, L.A. Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Res. 2013, 41, 7429–7437. [Google Scholar] [CrossRef] [Green Version]
- Didovyk, A.; Borek, B.; Tsimring, L.; Hasty, J. Transcriptional regulation with CRISPR-Cas9: Principles, advances, and applications. Curr. Opin. Biotechnol. 2016, 40, 177–184. [Google Scholar] [CrossRef] [Green Version]
- Gentzel, I.N.; Park, C.H.; Bellizzi, M.; Xiao, G.; Gadhave, K.R.; Murphree, C.; Yang, Q.; LaMantia, J.; Redinbaugh, M.G.; Balint-Kurti, P.; et al. A CRISPR/dCas9 toolkit for functional analysis of maize genes. Plant Methods 2020, 16, 133. [Google Scholar] [CrossRef]
- Li, Z.; Zhang, D.; Xiong, X.; Yan, B.; Xie, W.; Sheen, J.; Li, J.-F. A potent Cas9-derived gene activator for plant and mammalian cells. Nat. Plants 2017, 3, 930–936. [Google Scholar] [CrossRef] [Green Version]
- Vazquez-Vilar, M.; Bernabé-Orts, J.M.; Fernandez-Del-Carmen, A.; Ziarsolo, P.; Blanca, J.; Granell, A.; Orzaez, D. A modular toolbox for gRNA–Cas9 genome engineering in plants based on the GoldenBraid standard. Plant Methods 2016, 12, 10. [Google Scholar] [CrossRef] [Green Version]
- Lowder, L.G.; Zhang, D.; Baltes, N.J.; Paul, J.W.; Tang, X.; Zheng, X.; Voytas, D.; Hsieh, T.-F.; Zhang, Y.; Qi, Y. A CRISPR/Cas9 toolbox for multiplexed plant genome editing and transcriptional regulation. Plant Physiol. 2015, 169, 971–985. [Google Scholar] [CrossRef] [Green Version]
- Mahfouz, M.M.; Li, L.; Piatek, M.; Fang, X.; Mansour, H.; Bangarusamy, D.K.; Zhu, J.-K. Targeted transcriptional repression using a chimeric TALE-SRDX repressor protein. Plant Mol. Biol. 2011, 78, 311–321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, X.; Lowder, L.G.; Zhang, T.; Malzahn, A.A.; Zheng, X.; Voytas, D.F.; Zhong, Z.; Chen, Y.; Ren, Q.; Li, Q.; et al. A CRISPR–Cpf1 system for efficient genome editing and transcriptional repression in plants. Nat. Plants 2017, 3, 17018. [Google Scholar] [CrossRef] [PubMed]
- Lowder, L.G.; Zhou, J.; Zhang, Y.; Malzahn, A.; Zhong, Z.; Hsieh, T.-F.; Voytas, D.; Zhang, Y.; Qi, Y. Robust transcriptional activation in plants using multiplexed CRISPR-Act2.0 and mTALE-Act systems. Mol. Plant 2018, 11, 245–256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Papikian, A.; Liu, W.; Gallego-Bartolome, J.; Jacobsen, S.E. Site-specific manipulation of Arabidopsis loci using CRISPR-Cas9 SunTag systems. Nat. Commun. 2019, 10, 1–11. [Google Scholar] [CrossRef]
- Selma, S.; Bernabé-Orts, J.M.; Vazquez-Vilar, M.; Diego-Martin, B.; Ajenjo, M.; Garcia-Carpintero, V.; Granell, A.; Orzaez, D. Strong gene activation in plants with genome-wide specificity using a new orthogonal CRISPR/Cas9-based programmable transcriptional activator. Plant Biotechnol. J. 2019, 17, 1703–1705. [Google Scholar] [CrossRef] [Green Version]
- Zetsche, B.; Gootenberg, J.; Abudayyeh, O.O.; Slaymaker, I.M.; Makarova, K.S.; Essletzbichler, P.; Volz, S.E.; Joung, J.; van der Oost, J.; Regev, A.; et al. Cpf1 Is a Single RNA-guided endonuclease of a Class 2 CRISPR-Cas system. Cell 2015, 163, 759–771. [Google Scholar] [CrossRef] [Green Version]
- Fonfara, I.; Richter, H.; Bratovič, M.; Le Rhun, A.; Charpentier, E. The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature 2016, 532, 517–521. [Google Scholar] [CrossRef]
- Tanenbaum, M.E.; Gilbert, L.; Qi, L.S.; Weissman, J.S.; Vale, R.D. A Protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell 2014, 159, 635–646. [Google Scholar] [CrossRef] [Green Version]
- Konermann, S.; Brigham, M.; Trevino, A.E.; Joung, J.; Abudayyeh, O.O.; Barcena, C.; Hsu, P.; Habib, N.; Gootenberg, J.; Nishimasu, H.; et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 2014, 517, 583–588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, J.; Dempewolf, E.; Zhang, W.; Wang, Z.-Y. RNA-guided transcriptional activation via CRISPR/dCas9 mimics overexpression phenotypes in Arabidopsis. PLoS ONE 2017, 12, e0179410. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.E.; Neumann, M.; Duro, D.I.; Schmid, M. CRISPR-based tools for targeted transcriptional and epigenetic regulation in plants. PLoS ONE 2019, 14, e0222778. [Google Scholar] [CrossRef]
- Li, K.; Liu, Y.; Cao, H.; Zhang, Y.; Gu, Z.; Liu, X.; Yu, A.; Kaphle, P.; Dickerson, K.E.; Ni, M.; et al. Interrogation of enhancer function by enhancer-targeting CRISPR epigenetic editing. Nat. Commun. 2020, 11, 1–16. [Google Scholar] [CrossRef]
- Lowder, L.G.; Paul, J.W.; Qi, Y. Multiplexed transcriptional activation or repression in plants using CRISPR-dCas9-based systems. Plant Gene Regul. Netw. 2017, 1629, 167–184. [Google Scholar] [CrossRef]
- Azzeme, A.M.; Abdullah, S.N.A.; Aziz, M.A.; Wahab, P.E.M. Oil palm drought inducible DREB1 induced expression of DRE/CRT- and non-DRE/CRT-containing genes in lowland transgenic tomato under cold and PEG treatments. Plant Physiol. Biochem. 2017, 112, 129–151. [Google Scholar] [CrossRef] [PubMed]
- Baazim, H. RNA-Guided Transcriptional Regulation in Plants via dCas9 Chimeric Proteins. Ph.D. Thesis, King Abdullah University of Science and Technology Thuwal, Thuwal, Saudi Arabia, 2014. [Google Scholar]
- Bortesi, L.; Fischer, R. The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol. Adv. 2015, 33, 41–52. [Google Scholar] [CrossRef] [PubMed]
- Dreissig, S.; Schiml, S.; Schindele, P.; Weiss, O.; Rutten, T.; Schubert, V.; Gladilin, E.; Mette, M.F.; Puchta, H.; Houben, A. Live-cell CRISPR imaging in plants reveals dynamic telomere movements. Plant J. 2017, 91, 565–573. [Google Scholar] [CrossRef] [Green Version]
- Fujimoto, S.; Matsunaga, S. Visualization of chromatin loci with transiently expressed CRISPR/Cas9 in plants. Cytologia 2017, 82, 559–562. [Google Scholar] [CrossRef]
- Khosravi, S.; Schindele, P.; Gladilin, E.; Dunemann, F.; Rutten, T.; Puchta, H.; Houben, A. Application of aptamers improves CRISPR-based live imaging of plant telomeres. Front. Plant Sci. 2020, 11. [Google Scholar] [CrossRef]
- Gallego-Bartolome, J.; Gardiner, J.; Liu, W.; Papikian, A.; Ghoshal, B.; Kuo, H.Y.; Zhao, J.M.-C.; Segal, D.J.; Jacobsen, S.E. Targeted DNA demethylation of the Arabidopsis genome using the human TET1 catalytic domain. Proc. Natl. Acad. Sci. USA 2018, 115, E2125–E2134. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Yang, D.-L.; Huang, H.; Zhang, G.; He, L.; Pang, J.; Lozano-Durán, R.; Lang, Z.; Zhu, J.-K. Epigenetic memory marks determine epiallele stability at loci targeted by de novo DNA methylation. Nat. Plants 2020, 6, 661–674. [Google Scholar] [CrossRef]
- Morgan, S.L.; Mariano, N.C.; Bermudez, A.; Arruda, N.; Wu, F.; Luo, Y.; Shankar, G.; Jia, L.; Chen, H.; Hu, J.-F.; et al. Manipulation of nuclear architecture through CRISPR-mediated chromosomal looping. Nat. Commun. 2017, 8, 15993. [Google Scholar] [CrossRef] [Green Version]
- Joshi, R.; Wani, S.; Singh, B.; Bohra, A.; Dar, Z.; Lone, A.; Pareek, A.; Singla-Pareek, S.L. Transcription factors and plants response to drought stress: Current understanding and future directions. Front. Plant Sci. 2016, 7, 1029. [Google Scholar] [CrossRef] [Green Version]
- Paixão, J.F.R.; Gillet, F.-X.; Ribeiro, T.P.; Bournaud, C.; Lourenço-Tessutti, I.T.; Noriega, D.D.; De Melo, B.P.; De Almeida-Engler, J.; Grossi-De-Sa, M.F. Improved drought stress tolerance in Arabidopsis by CRISPR/dCas9 fusion with a Histone AcetylTransferase. Sci. Rep. 2019, 9, 1–9. [Google Scholar] [CrossRef] [Green Version]
- De Melo, B.P.; Lourenço-Tessutti, I.T.; Paixão, J.F.R.; Noriega, D.D.; Silva, M.C.M.; De Almeida-Engler, J.; Fontes, E.P.B.; Grossi-De-Sa, M.F. Transcriptional modulation of AREB-1 by CRISPRa improves plant physiological performance under severe water deficit. Sci. Rep. 2020, 10, 1–10. [Google Scholar] [CrossRef]
- Boualem, A.; Dogimont, C.; Bendahmane, A. The battle for survival between viruses and their host plants. Curr. Opin. Virol. 2016, 17, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Burgyán, J.; Havelda, Z. Viral suppressors of RNA silencing. Trends Plant Sci. 2011, 16, 265–272. [Google Scholar] [CrossRef] [PubMed]
- Abudayyeh, O.O.; Gootenberg, J.S.; Konermann, S.; Joung, J.; Slaymaker, I.M.; Cox, D.B.T.; Shmakov, S.; Makarova, K.S.; Semenova, E.; Minakhin, L.; et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 2016, 353, aaf5573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abudayyeh, O.O.; Gootenberg, J.S.; Essletzbichler, P.; Han, S.; Joung, J.; Belanto, J.J.; Verdine, V.; Cox, D.B.T.; Kellner, M.J.; Regev, A.; et al. RNA targeting with CRISPR–Cas13. Nature 2017, 550, 280–284. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.-Z.; Shi, M.; Holmes, E. Using metagenomics to characterize an expanding virosphere. Cell 2018, 172, 1168–1172. [Google Scholar] [CrossRef]
- Khan, Z.; Khan, S.H.; Ahmad, A.; Aslam, S.; Mubarik, M.S.; Khan, S. CRISPR:dCas9-mediated inhibition of replication of Begomoviruses. Int. J. Agric. Biol. 2019, 21, 711–718. [Google Scholar] [CrossRef]
- Aman, R.; Ali, Z.; Butt, H.; Mahas, A.; Aljedaani, F.; Khan, M.Z.; Ding, S.; Mahfouz, M. RNA virus interference via CRISPR/Cas13a system in plants. Genome Biol. 2018, 19, 1. [Google Scholar] [CrossRef] [PubMed]
- Gorlenko, C.L.; Kiselev, H.Y.; Budanova, E.V.; Zamyatnin, J.A.A.; Ikryannikova, L.N. Plant secondary metabolites in the battle of drugs and drug-resistant bacteria: New heroes or worse clones of antibiotics? Antibiotics 2020, 9, 170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, X.; Tang, K.; Li, P. Plant metabolic engineering strategies for the production of pharmaceutical terpenoids. Front. Plant Sci. 2016, 7, 1647. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reis, A.; Halper, S.M.; Vezeau, G.E.; Cetnar, D.P.; Hossain, A.; Clauer, P.R.; Salis, H.M. Simultaneous repression of multiple bacterial genes using nonrepetitive extra-long sgRNA arrays. Nat. Biotechnol. 2019, 37, 1294–1301. [Google Scholar] [CrossRef]
- Kim, S.K.; Han, G.H.; Seong, W.; Kim, H.; Kim, S.-W.; Lee, D.-H.; Lee, S.-G. CRISPR interference-guided balancing of a biosynthetic mevalonate pathway increases terpenoid production. Metab. Eng. 2016, 38, 228–240. [Google Scholar] [CrossRef]
- Jensen, E.D.; Ferreira, R.; Jakočiūnas, T.; Arsovska, D.; Zhang, J.; Ding, L.; Smith, J.D.; David, F.; Nielsen, J.; Jensen, M.K.; et al. Transcriptional reprogramming in yeast using dCas9 and combinatorial gRNA strategies. Microb. Cell Factories 2017, 16, 46. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.K.; Seong, W.; Han, G.H.; Lee, D.-H.; Lee, S.-G. CRISPR interference-guided multiplex repression of endogenous competing pathway genes for redirecting metabolic flux in Escherichia coli. Microb. Cell Factories 2017, 16, 188. [Google Scholar] [CrossRef] [Green Version]
- Schwartz, C.; Curtis, N.; Löbs, A.-K.; Wheeldon, I. Multiplexed CRISPR activation of cryptic sugar metabolism enables Yarrowia lipolytica growth on cellobiose. Biotechnol. J. 2018, 13, e1700584. [Google Scholar] [CrossRef] [PubMed]
- Puchta, H. Applying CRISPR/Cas for genome engineering in plants: The best is yet to come. Curr. Opin. Plant Biol. 2017, 36, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Hilton, I.B.; D’Ippolito, A.M.; Vockley, C.M.; Thakore, P.I.; Crawford, G.E.; Reddy, T.E.; Gersbach, C.A. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat. Biotechnol. 2015, 33, 510–517. [Google Scholar] [CrossRef] [Green Version]
- Kawall, K.; Cotter, J.; Then, C. Broadening the GMO risk assessment in the EU for genome editing technologies in agriculture. Environ. Sci. Eur. 2020, 32, 106. [Google Scholar] [CrossRef]
- Concordet, J.-P.; Haeussler, M. CRISPOR: Intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res. 2018, 46, W242–W245. [Google Scholar] [CrossRef] [PubMed]
- Stemmer, M.; Thumberger, T.; Keyer, M.D.S.; Wittbrodt, J.; Mateo, J.L. CCTop: An intuitive, flexible and reliable CRISPR/Cas9 target prediction tool. PLoS ONE 2015, 10, e0124633. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, H.; Guo, J.; Wang, T.; Zhang, C.; Xing, X.-H. Guide-target mismatch effects on dCas9–sgRNA binding activity in living bacterial cells. Nucleic Acids Res. 2021, 49, 1263–1277. [Google Scholar] [CrossRef] [PubMed]
- Larson, M.H.; Gilbert, L.; Wang, X.; Lim, W.A.; Weissman, J.S.; Qi, L.S. CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat. Protoc. 2013, 8, 2180–2196. [Google Scholar] [CrossRef] [Green Version]
- Choudhary, E.; Lunge, A.; Agarwal, N. Strategies of genome editing in mycobacteria: Achievements and challenges. Tuberculosis 2016, 98, 132–138. [Google Scholar] [CrossRef]
- Eckerstorfer, M.F.; Dolezel, M.; Heissenberger, A.; Miklau, M.; Reichenbecher, W.; Steinbrecher, R.A.; Waßmann, F. An EU perspective on biosafety considerations for plants developed by Genome editing and other new genetic modification techniques (nGMs). Front. Bioeng. Biotechnol. 2019, 7, 31. [Google Scholar] [CrossRef] [Green Version]
- Agapito-Tenfen, S.Z.; Okoli, A.; Bernstein, M.J.; Wikmark, O.-G.; Myhr, A.I. Revisiting risk governance of GM plants: The need to consider new and emerging gene-editing techniques. Front. Plant Sci. 2018, 9, 1874. [Google Scholar] [CrossRef] [Green Version]
- Singh, O.V.; Ghai, S.; Paul, D.; Jain, R.K. Genetically modified crops: Success, safety assessment, and public concern. Appl. Microbiol. Biotechnol. 2006, 71, 598–607. [Google Scholar] [CrossRef]
- Cui, K.; Shoemaker, S.P. Public perception of genetically-modified (GM) food: A Nationwide Chinese Consumer Study. npj Sci. Food 2018, 2, 1–8. [Google Scholar] [CrossRef]
- State Council of China. Administrative Regulations on Safety of Agricultural GMOs. 2001, supra Note 7, Art. 11. Available online: http://www.moa.gov.cn/ztzl/zjyqwgz/zcfg/201107/t20110711_2049870.htm (accessed on 24 January 2020). (In Chinese)
- European Parliament. Directive 2001/18/EC of the European Parliament and of the Council of 12 March 2001 on the Deliberate Release into the Environment of Genetically Modified Organisms and Repealing Council Directive 90/220/EEC—Commission Declaration. Off. J. 2001, 106, 1–39. [Google Scholar]
- Globus, R.; Qimron, U. A technological and regulatory outlook on CRISPR crop editing. J. Cell. Biochem. 2017, 119, 1291–1298. [Google Scholar] [CrossRef] [PubMed]
- Waltz, E. Gene-edited CRISPR mushroom escapes US regulation. Nature 2016, 532, 293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- USDA. Petitions for Determination of Nonregulated Status; USDA: Washington, DC, USA, 2020. [Google Scholar]




| CRISPR/dCas9 | Modification | Plant Species | Target Gene | Repression (%)/Activation (Fold-Change) | References |
|---|---|---|---|---|---|
| Transcriptional suppression | dCas9-SRDX | Nicotiana benthamiana | pNOS::LUC reporter | 33 | [95] |
| dCas9-BRD | N. benthamiana | pNOS::LUC reporter | 60 | [109] | |
| dCas9-3xSRDX | Arabidopsis | CFTS64 | 60 | [110] | |
| dCas9-TALE-SRDX | Arabidopsis | RD29-LUC (1) | - | [111] | |
| dLbCpf1-SRDX | Arabidopsis | miR159B | 90 | [112] | |
| dAsCpf1-SRDX | Arabidopsis | miR159B | 90 | [112] | |
| Transcriptional activation | dCas9VP64 | Oryza sativa | Os03g01240 | 2.0 | [113] |
| dCas9-TV | O. sativa | OsER1 | 62.0 | [108] | |
| dCas9VP64+MS2-VP64 | O. sativa | Os04g39780 | 4.0 | [113] | |
| dCas9-VP64 | Arabidopsis | pWRKY::luciferase | 6.7 | [108] | |
| dCas9-MCP-TV | Arabidopsis | AtWRKY | 11.7 | [108] | |
| dCas9-SunTag | Arabidopsis | AtCLAVATA3 | 100.0 | [114] | |
| dCas9:SunTag-EDLL | N. benthamiana | pNOS::luciferase | 3.0 | [115] | |
| dCas9-VP64 | N. benthamiana | pNOS::luciferase | 3.0 | [95] | |
| dCas9-EDLL | N. benthamiana | NbPDS | 3.4 | [95] |
| Application | Plant Species | Modification | Reference |
|---|---|---|---|
| Live cell chromatin imaging | N. benthamiana | dCas9-eGFP | [127] |
| dCas9-FP | [128] | ||
| dCas9-MS2-mRuby2 | [129] | ||
| Transcriptional regulation | Arabidopsis | dCas9-MCP-TV | [108] |
| dCas9-3xSRDX | [110] | ||
| dAsCpf1-SRDX | [112] | ||
| O. sativa | LUC/dCas9-TV | [123] | |
| dCas9VP64+ MS2-VP64 | [113] | ||
| dCas9-TV-6 × His | [108] | ||
| N. benthamiana | dCas9-VP128 | [95] | |
| dCas9-EDLL | [95] | ||
| Epigenetic manipulation | Arabidopsis | dCas9-MS2 | [121] |
| dCas9-TET1cd | [130,131] | ||
| dCas9-SunTag | [114] | ||
| Chromatin topology | Arabidopsis | dCas9-PYL1 | [132] |
| dCas9-ABI1 | [89] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karlson, C.K.S.; Mohd-Noor, S.N.; Nolte, N.; Tan, B.C. CRISPR/dCas9-Based Systems: Mechanisms and Applications in Plant Sciences. Plants 2021, 10, 2055. https://0-doi-org.brum.beds.ac.uk/10.3390/plants10102055
Karlson CKS, Mohd-Noor SN, Nolte N, Tan BC. CRISPR/dCas9-Based Systems: Mechanisms and Applications in Plant Sciences. Plants. 2021; 10(10):2055. https://0-doi-org.brum.beds.ac.uk/10.3390/plants10102055
Chicago/Turabian StyleKarlson, Chou Khai Soong, Siti Nurfadhlina Mohd-Noor, Nadja Nolte, and Boon Chin Tan. 2021. "CRISPR/dCas9-Based Systems: Mechanisms and Applications in Plant Sciences" Plants 10, no. 10: 2055. https://0-doi-org.brum.beds.ac.uk/10.3390/plants10102055