Improving the Efficiency of Adventitious Shoot Induction and Somatic Embryogenesis via Modification of WUSCHEL and LEAFY COTYLEDON 1
Abstract
:1. Introduction
2. Results
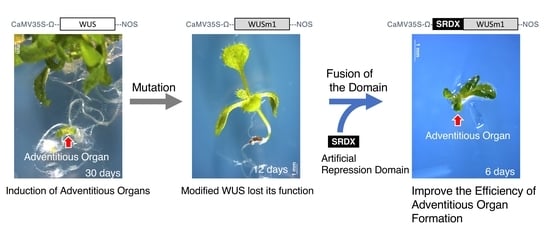
2.1. The WUS-Box Is the Main Functional Domain for Adventitious Organ Formation
2.2. Fusion with an Artificial Repression Domain Enhances the Ability of WUS to Induce Adventitious Organ Formation
2.3. Fusion of an Artificial Activation Domain Enhances the Ability of LEC1 to Induce Callus Formation
3. Discussion
4. Materials and Methods
4.1. Growth and Transformation of Plants
4.2. Construction of Plasmids
4.3. Microscopy Observations
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Zuo, J.; Niu, Q.W.; Frugis, G.; Chua, N.H. The WUSCHEL gene promotes vegetative-to-embryonic transition in Arabidopsis. Plant J. 2002, 30, 349–359. [Google Scholar] [CrossRef] [PubMed]
- Lotan, T.; Ohto, M.; Yee, K.M.; West, M.A.; Lo, R.; Kwong, R.W.; Yamagishi, K.; Fischer, R.L.; Goldberg, R.B.; Harada, J.J. Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 1998, 93, 1195–1205. [Google Scholar] [CrossRef] [Green Version]
- Ikeda, M.; Mitsuda, N.; Ohme-Takagi, M. Arabidopsis WUSCHEL is a bifunctional transcription factor that acts as a repressor in stem cell regulation and as an activator in floral patterning. Plant Cell 2009, 21, 3493–3505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laux, T.; Mayer, K.F.; Berger, J.; Jürgens, G. The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 1996, 122, 87–96. [Google Scholar]
- Mayer, K.F.; Schoof, H.; Haecker, A.; Lenhard, M.; Jürgens, G.; Laux, T. Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 1998, 95, 805–815. [Google Scholar] [CrossRef] [Green Version]
- Gallois, J.L.; Nora, F.R.; Mizukami, Y.; Sablowski, R. WUSCHEL induces shoot stem cell activity and developmental plasticity in the root meristem. Genes Dev. 2004, 18, 375–380. [Google Scholar] [CrossRef] [Green Version]
- Rashid, S.Z.; Yamaji, N.; Kyo, M. Shoot formation from root tip region: A developmental alteration by WUS in transgenic tobacco. Plant Cell Rep. 2007, 26, 1449–1455. [Google Scholar] [CrossRef]
- Arroyo-Herrera, A.; Gonzalez, A.K.; Moo, R.C.; Quiroz-Figueroa, F.R.; Loyola-Vargas, V.M.; Rodriguez-Zapata, L.C.; Dhondt, C.B.; Suarez-Solis, V.M.; Castano, E. Expression of WUSCHEL in Coffea canephora causes ectopic morphogenesis and increases somatic embryogenesis. Plant Cell Tissue Organ Cult. 2008, 94, 171–180. [Google Scholar] [CrossRef]
- Bouchabké-Coussa, O.; Obellianne, M.; Linderme, D.; Montes, E.; Maia-Grondard, A.; Vilaine, F.; Pannetier, C. Wuschel overexpression promotes somatic embryogenesis and induces organogenesis in cotton (Gossypium hirsutum L.) tissues cultured in vitro. Plant Cell Rep. 2013, 32, 675–686. [Google Scholar] [CrossRef]
- Zheng, W.; Zhang, X.; Yang, Z.; Wu, J.; Li, F.; Duan, L.; Liu, C.; Lu, L.; Zhang, C. AtWuschel promotes formation of the embryogenic callus in Gossypium hirsutum. PLoS ONE 2014, 9, e87502. [Google Scholar] [CrossRef] [Green Version]
- Lowe, K.; Wu, E.; Wang, N.; Hoerster, G.; Hastings, C.; Cho, M.J.; Scelonge, C.; Lenderts, B.; Chamberlin, M.; Cushatt, J.; et al. Morphogenic Regulators Baby boom and Wuschel Improve Monocot Transformation. Plant Cell 2016, 28, 1998–2015. [Google Scholar] [CrossRef] [Green Version]
- Lowe, K.; La Rota, M.; Hoerster, G.; Hastings, C.; Wang, N.; Chamberlin, M.; Wu, E.; Jones, T.; Gordon-Kamm, W. Rapid genotype “independent” Zea mays L. (maize) transformation via direct somatic embryogenesis. Cell Dev. Biol. Plant 2018, 54, 240–252. [Google Scholar] [CrossRef] [Green Version]
- Niu, D.; He, Y. Leafy Cotyledons: Old genes with new roles beyond seed development. F1000Res 2019, 8. [Google Scholar] [CrossRef] [PubMed]
- Pelletier, J.M.; Kwong, R.W.; Park, S.; Le, B.H.; Baden, R.; Cagliari, A.; Hashimoto, M.; Munoz, M.D.; Fischer, R.L.; Goldberg, R.B.; et al. LEC1 sequentially regulates the transcription of genes involved in diverse developmental processes during seed development. Proc. Natl. Acad. Sci. USA 2017, 114, E6710–E6719. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.; Wong, L.; Meng, L.; Lemaux, P.G. Similarity of expression patterns of knotted1 and ZmLEC1 during somatic and zygotic embryogenesis in maize (Zea mays L.). Planta 2002, 215, 191–194. [Google Scholar] [CrossRef] [PubMed]
- Ikeda-Iwai, M.; Satoh, S.; Kamada, H. Establishment of a reproducible tissue culture system for the induction of Arabidopsis somatic embryos. J. Exp. Bot. 2002, 53, 1575–1580. [Google Scholar] [CrossRef] [Green Version]
- Yazawa, K.; Takahata, K.; Kamada, H. Isolation of the gene encoding Carrot leafy cotyledon1 and expression analysis during somatic and zygotic embryogenesis. Plant Physiol. Biochem. 2004, 42, 215–223. [Google Scholar] [CrossRef] [PubMed]
- Rupps, A.; Raschke, J.; Rümmler, M.; Linke, B.; Zoglauer, K. Identification of putative homologs of Larix decidua to babyboom (bbm), leafy cotyledon1 (lec1), wuschel-related homeobox2 (wox2) and somatic embryogenesis receptor-like KINASE (SERK) during somatic embryogenesis. Planta 2016, 243, 473–488. [Google Scholar] [CrossRef]
- Brand, A.; Quimbaya, M.; Tohme, J.; Chavarriaga-Aguirre, P. Arabidopsis LEC1 and LEC2 Orthologous Genes Are Key Regulators of Somatic Embryogenesis in Cassava. Front. Plant Sci. 2019, 10, 673. [Google Scholar] [CrossRef]
- Guo, F.; Liu, C.; Xia, H.; Bi, Y.; Zhao, C.; Zhao, S.; Hou, L.; Li, F.; Wang, X. Induced expression of AtLEC1 and AtLEC2 differentially promotes somatic embryogenesis in transgenic tobacco plants. PLoS ONE 2013, 8, e71714. [Google Scholar] [CrossRef] [Green Version]
- Hiratsu, K.; Matsui, K.; Koyama, T.; Ohme-Takagi, M. Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 2003, 34, 733–739. [Google Scholar] [CrossRef] [PubMed]
- Triezenberg, S.J.; Kingsbury, R.C.; McKnight, S.L. Functional dissection of VP16, the trans-activator of herpes simplex virus immediate early gene expression. Genes Dev. 1988, 2, 718–729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagaya, S.; Kawamura, K.; Shinmyo, A.; Kato, K. The HSP terminator of Arabidopsis thaliana increases gene expression in plant cells. Plant Cell Physiol. 2010, 51, 328–332. [Google Scholar] [CrossRef] [Green Version]
- Lenhard, M.; Jürgens, G.; Laux, T. The wuschel and shootmeristemless genes fulfil complementary roles in Arabidopsis shoot meristem regulation. Development 2002, 129, 3195–3206. [Google Scholar]
- Gallois, J.L.; Woodward, C.; Reddy, G.V.; Sablowski, R. Combined shoot meristemless and wuschel trigger ectopic organogenesis in Arabidopsis. Development 2002, 129, 3207–3217. [Google Scholar]
- Banno, H.; Ikeda, Y.; Niu, Q.W.; Chua, N.H. Overexpression of Arabidopsis ESR1 induces initiation of shoot regeneration. Plant Cell 2001, 13, 2609–2618. [Google Scholar] [CrossRef]
- Ikeda, Y.; Banno, H.; Niu, Q.W.; Howell, S.H.; Chua, N.H. The enhancer of shoot regeneration 2 gene in Arabidopsis regulates cup-shaped cotyledon 1 at the transcriptional level and controls cotyledon development. Plant Cell Physiol. 2006, 47, 1443–1456. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boutilier, K.; Offringa, R.; Sharma, V.K.; Kieft, H.; Ouellet, T.; Zhang, L.; Hattori, J.; Liu, C.M.; van Lammeren, A.A.; Miki, B.L.; et al. Ectopic expression of baby boom triggers a conversion from vegetative to embryonic growth. Plant Cell 2002, 14, 1737–1749. [Google Scholar] [CrossRef] [Green Version]
- Horstman, A.; Li, M.; Heidmann, I.; Weemen, M.; Chen, B.; Muino, J.M.; Angenent, G.C.; Boutilier, K. The baby boom Transcription Factor Activates the LEC1-ABI3-FUS3-LEC2 Network to Induce Somatic Embryogenesis. Plant Physiol. 2017, 175, 848–857. [Google Scholar] [CrossRef] [Green Version]
- Stone, S.L.; Braybrook, S.A.; Paula, S.L.; Kwong, L.W.; Meuser, J.; Pelletier, J.; Hsieh, T.F.; Fischer, R.L.; Goldberg, R.B.; Harada, J.J. Arabidopsis leafy cotyledon2 induces maturation traits and auxin activity: Implications for somatic embryogenesis. Proc. Natl. Acad. Sci. USA 2008, 105, 3151–3156. [Google Scholar] [CrossRef] [Green Version]
- Gaj, M.D.; Zhang, S.; Harada, J.J.; Lemaux, P.G. Leafy cotyledon genes are essential for induction of somatic embryogenesis of Arabidopsis. Planta 2005, 222, 977–988. [Google Scholar] [CrossRef] [PubMed]
- Wójcikowska, B.; Jaskóła, K.; Gąsiorek, P.; Meus, M.; Nowak, K.; Gaj, M.D. Leafy Cotyledon2 (Lec2) promotes embryogenic induction in somatic tissues of Arabidopsis, via YUCCA-mediated auxin biosynthesis. Planta 2013, 238, 425–440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mookkan, M.; Nelson-Vasilchik, K.; Hague, J.; Zhang, Z.J.; Kausch, A.P. Selectable marker independent transformation of recalcitrant maize inbred B73 and sorghum P898012 mediated by morphogenic regulators baby boom and Wuschel2. Plant Cell Rep. 2017, 36, 1477–1491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oshima, Y.; Mitsuda, N.; Nakata, M.; Nakagawa, T.; Nagaya, S.; Kato, K.; Ohme-Takagi, M. Novel vector systems to accelerate functional analysis of transcription factors using chimeric repressor gene-silencing technology (CRES-T). Plant Biotechnol. 2011, 28, 201–210. [Google Scholar] [CrossRef] [Green Version]
- Mitsuda, N.; Hiratsu, K.; Todaka, D.; Nakashima, K.; Yamaguchi-Shinozaki, K.; Ohme-Takagi, M. Efficient production of male and female sterile plants by expression of a chimeric repressor in Arabidopsis and rice. Plant Biotechnol. J. 2006, 4, 325–332. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, S.; Kigoshi, K.; Mitsuda, N.; Suzuki, K.; Ohme-Takagi, M. VP16 fusion efficiently reveals the function of transcriptional repressors in Arabidopsis. Plant Biotechnol. 2014, 1, 123–132. [Google Scholar] [CrossRef] [Green Version]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ikeda, M.; Takahashi, M.; Fujiwara, S.; Mitsuda, N.; Ohme-Takagi, M. Improving the Efficiency of Adventitious Shoot Induction and Somatic Embryogenesis via Modification of WUSCHEL and LEAFY COTYLEDON 1. Plants 2020, 9, 1434. https://0-doi-org.brum.beds.ac.uk/10.3390/plants9111434
Ikeda M, Takahashi M, Fujiwara S, Mitsuda N, Ohme-Takagi M. Improving the Efficiency of Adventitious Shoot Induction and Somatic Embryogenesis via Modification of WUSCHEL and LEAFY COTYLEDON 1. Plants. 2020; 9(11):1434. https://0-doi-org.brum.beds.ac.uk/10.3390/plants9111434
Chicago/Turabian StyleIkeda, Miho, Mikiya Takahashi, Sumire Fujiwara, Nobutaka Mitsuda, and Masaru Ohme-Takagi. 2020. "Improving the Efficiency of Adventitious Shoot Induction and Somatic Embryogenesis via Modification of WUSCHEL and LEAFY COTYLEDON 1" Plants 9, no. 11: 1434. https://0-doi-org.brum.beds.ac.uk/10.3390/plants9111434