Colorimetric Sensing of Amoxicillin Facilitated by Molecularly Imprinted Polymers
Abstract
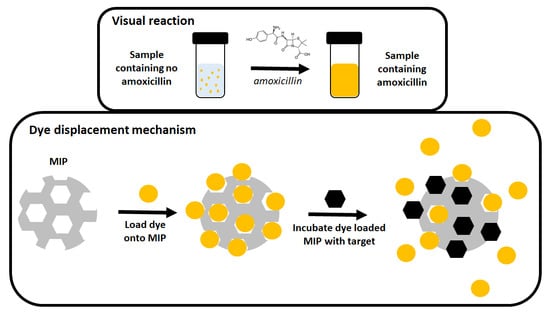
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Equipment
2.2. Generalised Monolithic Bulk Imprinting Method
2.3. Generalized MIP Synthesis by Emulsion Polymerization
2.4. Generalised Batch Rebinding Experiment
2.5. Preloading of Dye to MIPs
2.6. Dose Response of Dye Displacement
3. Results
3.1. Analysis of Bulk MIPs
3.2. Emulsion Particle Size Analysis
3.3. Emulsion MIP Binding Analysis
3.4. Selection of Dye
3.5. Dye Displacement
4. Discussion
4.1. Bulk Polymerization MIP Analysis
4.2. Emulsion MIP Structure
4.3. Binding of Emulsion MIP
4.4. Selection of a Dye for Displacement
4.5. Dye Displacement Assay
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A

References
- Kummerer, K. Significance of antibiotics in the environment. J. Antimicrob. Chemother. 2003, 52, 5–7. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Liu, B.; Zhang, X.; Wang, J.; Gao, S. The distribution of veterinary antibiotics in the river system in a livestock-producing region and interactions between different phases. Environ. Sci. Pollut. Res. 2016, 23, 16542–16551. [Google Scholar] [CrossRef]
- Zhi, S.; Shen, S.; Zhou, J.; Ding, G.; Zhang, K. Systematic analysis of occurence, density and ecological risks of 45 veterinary antibiotics: Focused on family livestock farms in Erthai Lake basin, Yunnan, China. Environ. Pollut. 2020, 267, 115539. [Google Scholar] [CrossRef]
- Wang, L.; Yang, H.; Zhang, C.; Mo, Y.; Lu, X. Determination of oxytetracycline, tetracycline and chloramphenicol antibiotics in animal feeds using subcritical water extraction and high performance liquid chromatography. Anal. Chim. Acta 2008, 619, 54–58. [Google Scholar] [CrossRef] [PubMed]
- Daghrir, R.; Drogui, P. Tetracycline antibiotics in the environment: A review. Environ. Chem. Lett. 2013, 11, 209–227. [Google Scholar] [CrossRef]
- Michael, I.; Rizzo, L.; McArdell, C.S.; Manaia, C.M.; Merlin, C.; Schwartz, T.; Dagot, C.; Fatta-Kassinos, D. Urban wastewater treatment plants as hotspots for the release of antibiotics in the environment: A review. Water Res. 2013, 47, 957–995. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kantiani, L.; Farre, M.; Barcelo, D.; Barcelo, D. Analytical methodologies for the detection of β-lactam antibiotics in milk and feed samples. TrAC 2009, 28, 729–744. [Google Scholar] [CrossRef]
- Jing, T.; Wang, Y.; Dai, Q.; Xia, H.; Niu, J.; Hao, Q.; Mei, S.; Zhou, Y. Preperation of mixed-templates molecularly imprinted polymers and investigation of the recognition ability for tetracycline antibiotics. Biosens. Bioelectron. 2010, 25, 2218–2224. [Google Scholar] [CrossRef] [PubMed]
- Luo, Q.; Yu, N.; Shi, C.; Wang, X.; Wu, J. Surface plasmon resonance sensor for antibiotics detection based on photo-initiated polymerization molecularly imprinted array. Talanta 2016, 161, 797–803. [Google Scholar] [CrossRef] [PubMed]
- Naklua, W.; Suedee, R.; Lieberzeit, P.A. Dopaminergic receptor-ligand binding assay based on molecularly imprinted polymers on quartz crystal microbalance sensors. Biosens. Bioelectron. 2016, 81, 117–124. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Tran, T.M.; Elhaj, A.A.A.; Torsetnes, S.B.; Jensen, O.N.; Sellergren, B.; Irgum, K. Molecularly Imprinted Porous Monolithic Materials From Melamine-Formaldehyde for Selective Trapping of Phosphopeptides. Anal. Chem. 2017, 89, 9491–9501. [Google Scholar] [CrossRef] [PubMed]
- Lowdon, J.W.; Alkirkit, S.M.O.; Mewis, R.E.; Fulton, D.; Banks, C.E.; Sutcliffe, O.B.; Peeters, M. Engineering molecularly imprinted polymers (MIPs) for the selective extraction and quantification of the novel psychoactive substance (NPS) methoxphenidine and its regioisomers. Analyst 2018, 143, 2081–2089. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, J.; Bai, J.; Peng, Y.; Qie, Z.; Zhao, Y.; Ning, B.; Xiao, D.; Gao, Z. A core-shell-structured molecularly imprinted polymer on upconverting nanoparticles for selective and sensitive fluorescence sensing sulfamethazine. Analyst 2015, 140, 5301–5307. [Google Scholar] [CrossRef] [PubMed]
- Qu, Y.; Qin, L.; Lui, X.; Yang, Y. Reasonable design and sifting of microporous carbon nanosphere-based surface molecularly imprinted polymer for selective removal of phenol from wastewater. Chemosphere 2020, 251, 126376. [Google Scholar] [CrossRef]
- Pisarev, O.A.; Polyakova, I.V. Molecularly imprinted polymers based on methacrylic acid and ethyleneglycol dimethacrylate for L-lysine recognition. React. Funct. Polym. 2018, 130, 98–110. [Google Scholar] [CrossRef]
- Cantarella, M.; Carroccio, S.C.; Dattilo, S.; Avolio, R.; Castaldo, R.; Puglisi, C.; Privitera, V. Molecularly imprinted polymer for the selective absorption of diclofenac from contaminated water. Chem. Eng. J. 2019, 367, 180–188. [Google Scholar] [CrossRef]
- Vandenryt, T.; van Grinsven, B.; Eersels, K.; Cornelis, P.; Kholwadia, S.; Cleij, T.J.; Thoelen, R.; De Ceuninck, W.; Wagner, P. Single-shot detection of neurotransmitters in whole-blood samples by means of the heat-transfer method in combination with synthetic receptors. Sensors 2017, 17, 2701. [Google Scholar] [CrossRef] [Green Version]
- Eersels, K.; Dilien, H.; Lowdon, J.W.; Redeker, E.S.; Rogosic, R.; Heidt, B.; Peeters, M.; Cornelis, P.; Lux, P.; Reutelingsperger, C.P.; et al. A novel biomimetic tool for assessing vitamin K status based on molecularly imprinted polymers. Nutrients 2018, 10, 751. [Google Scholar] [CrossRef] [Green Version]
- Casadio, S.; Lowdon, J.W.; Betlem, K.; Ueta, J.T.; Foster, C.W.; Cleij, T.J.; Grinsven, B.; Sutcliffe, O.B.; Banks, C.E.; Peeters, M. Development of a novel flexible polymer-based biosensor for the thermal detection of noradrenaline in aqueous solutions. Chem. Eng. J. 2017, 315, 459–468. [Google Scholar] [CrossRef]
- Lowdon, J.W.; Dilien, H.; Singla, P.; Peeters, M.; Cleij, T.J.; van Grinsven, B.; Eersels, K. MIPs for commercial application in low-cost sensors and assays—An overview of the current status quo. Sens. Actuators B Chem. 2020, 325, 128973. [Google Scholar] [CrossRef]
- McNiven, S.; Kato, M.; Levi, R.; Yano, K.; Karube, I. Chloramphenicol sensor based on an in situ imprinted polymer. Anal. Chim. Acta 1998, 365, 69–74. [Google Scholar] [CrossRef]
- Greene, N.T.; Shimizu, K.D. Colorimetric molecularly imprinted polymer sensor array using dye displacement. J. Am. Chem. Soc. 2005, 127, 5695–5700. [Google Scholar] [CrossRef]
- Mattsson, L.; Xu, J.; Preininger, C.; Tse Sum Bui, B.; Haupt, K. Competitive fluorescent pseudo-immunoassay exploiting molecularly imprinted polymers for the detection of biogenic amines in fish matrix. Talanta 2018, 181, 190–196. [Google Scholar] [CrossRef]
- Li, C.; Ngai, M.H.; Reddy, K.K.; Leong, S.C.Y.; Tong, Y.W.; Chai, C.L.L. A fluorescence-displacement assay using molecularly imprinted polymers for the visual, rapid, and sensitive detection of the algal metabolites, geosim and 2-methylisoborneol. Anal. Chim. Acta 2019, 1066, 121–130. [Google Scholar] [CrossRef]
- Silverio, O.V.; So, R.C.; Elnar, K.J.S.; Malapit, C.A.; Nepomuceno, M.C.M. Development of dieldrin, endosulfan, and hexachlorobenzene-imprinted polymers for dye-displacement array sensing. J. Appl. Polym. Sci. 2017, 134, 44401. [Google Scholar] [CrossRef]
- Lowdon, J.W.; Eersels, K.; Rogosic, R.; Heidt, B.; Diliën, H.; Redeker, E.S.; Peeters, M.; van Grinsiven, B.; Cleij, T.J. Substrate Displacement Colorimetry for the Detection of Diarylethylamines. Sens. Actuators B Chem. 2019, 282, 137–144. [Google Scholar] [CrossRef]
- Lowdon, J.W.; Eersels, K.; Arreguin-Campos, R.; Caldara, M.; Heidt, B.; Rogosic, R.; Jimenez-Monroy, K.L.; Cleij, T.J.; Diliën, H.; van Grinsven, B. A molecularly imprinted polymer-based dye displacement assay for the rapid visual detection of amphetamine in urine. Molecules 2020, 25, 5222. [Google Scholar] [CrossRef] [PubMed]
- Romano, A.; Valluzzi, R.L.; Caruso, C.; Zaffiro, A.; Quaratino, D.; Gaeta, F. Tolerability of cefazolin and ceftibuten in patients with IgE-mediated aminopenicillin allergy. J. Allergy Clin. Immunol. Pract. 2020, 8, 1989–1993. [Google Scholar] [CrossRef] [PubMed]
- Mollmann, U.; Heinisch, L.; Bauernfeind, A.; Kohler, T.; Ankel-Fuchs, D. Siderophores as drug delivery agents: Application of the “Trojan Horse” strategy. BioMetals 2009, 22, 615–624. [Google Scholar] [CrossRef] [PubMed]
- Kushwaha, A.; Srivastava, J.; Singh, A.K.; Anand, R.; Raghuwanshi, R.; Rai, T.; Singh, M. Epitope imprinting of Mycobacterium leprae bacteria via molecularly imprinted nanoparticles using multiple monomers approach. Biosens. Bioelectron. 2019, 145, 111698. [Google Scholar] [CrossRef]
- Duan, F.; Chen, C.; Zhao, X.; Yang, Y.; Liu, X.; Qin, Y. Water-compatible surface molecularly imprinted polymers with synergy of bi-functional monomers for enhanced selective adsorption of bisphenol A from aqueous solution. Environ. Sci. Nano 2016, 3, 213–222. [Google Scholar] [CrossRef]
- Wloch, M.; Datta, J. Chapter Two—Synthesis and polymerization techniques of molecularly imprinted polymers. Compr. Anal. Chem. 2019, 86, 17–40. [Google Scholar]
- Erturk, G.; Mattiasson, B. Molecular imprinting techniques used for the preparation of biosensors. Sensors 2017, 17, 288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Villa, C.C.; Sanchez, L.; Valencia, G.A.; Ahmed, S.; Gutierrez, T.J. Molecularly imprinted polymers for food applications: A review. Trends Food Sci. Technol. 2021, 111, 642–669. [Google Scholar] [CrossRef]
- Ou, H.; Chen, Q.; Pan, J.; Zhang, Y.; Huang, Y.; Qi, X. Selective removal of erythromycin by magnetic imprinted polymers synthesized from chitosan-stabilized pickering emulsion. J. Hazard. Mat. 2015, 289, 28–37. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Li, L.; Hang, H.; Wu, R.; Dai, X.; Shi, W.; Yan, Y. Fabrication and evaluation of magnetic/hollow double-shelled imprinted sorbents formed by pickering emulsion polymerization. Langmuir 2013, 29, 8170–8178. [Google Scholar] [CrossRef] [PubMed]
- Lim, K.F.; Holdsworth, C.I. Effect of formulation on the binding efficiency and selectivity of precipitation molecularly imprinted polymers. Molecules 2018, 23, 2996. [Google Scholar] [CrossRef] [Green Version]
- Pardeshi, S.; Singh, S.K. Precipitation polymerization: A versitile tool for preparing molecularly imprinted polymer beads for chromatography applications. RSC Adv. 2016, 6, 23525–23536. [Google Scholar] [CrossRef]
- Canfarotta, F.; Poma, A.; Guerreiro, A.; Piletsky, S. Solid-phase synthesis of molecularly imprinted nanoparticles. Nat. Protoc. 2016, 11, 443–455. [Google Scholar] [CrossRef] [PubMed]
- Refaat, D.; Aggour, M.G.; Farghali, A.A.; Mahajan, R.; Wiklander, J.G.; Nicholls, I.A.; Piletsky, S.A. Strategies for molecular imprinting and the evolution of MIP nanoparticles as plastic antibodies—Synthesis and applications. Int. J. Mol. Sci. 2019, 20, 6304. [Google Scholar] [CrossRef] [Green Version]
- Orowitz, T.E.; Sombo, P.P.A.A.A.; Rahayu, D.; Hasanah, A.N. Microsphere polymers in molecular imprinting: Current and future perspectives. Molecules 2020, 25, 3256. [Google Scholar] [CrossRef] [PubMed]
- Jamieson, O.; Soares, T.C.C.; Faria, B.A.; Hudson, A.; Mecozzi, F.; Rowley-Neale, S.J.; Banks, C.E.; Gruber, J.; Novabovic, K.; Peeters, M.; et al. Screen printed electrode based detection systems for the antibiotic amoxicillin in aqueous samples utilizing molecularly imprinted polymers as synthetic receptors. Chemosensors 2020, 8, 5. [Google Scholar] [CrossRef] [Green Version]
- Rolinson, G.N. Laboratory evaluation of amoxicillin. J. Intect. Dis. 1974, 129, S139–S145. [Google Scholar] [CrossRef] [PubMed]
- Katz, A.; Davis, M.E. Molecular imprinting of bulk, microporous silica. Nature 2000, 403, 286–289. [Google Scholar] [CrossRef]
- Alabdullah, S.S.M.; Ismail, H.K.; Ryder, K.S.; Abbott, A.P. Evidence supporting an emulsion polymerization mechanism for the formation of polyaniline. Electochim. Acta 2020, 354, 136737. [Google Scholar] [CrossRef]
- Wang, D.; Zhu, J.; Yao, Q.; Wilkie, C.A. A comparison of various methods for the preparation of polystyrene and poly(methyl methacrylate) clay nanocomposites. Chem. Mater. 2002, 14, 3837–3843. [Google Scholar] [CrossRef]
- Silva, R.C.S.; Santos, M.N.; Pires, B.C.; Dinali, L.A.F.; Suquila, F.A.C.; Tarley, C.R.T.; Borges, K.B. Assessment of surfactants on performance of molecularly imprinted polymer toward absorption of pharmaceutical. J. Environ. Chem. Eng. 2019, 7, 103037. [Google Scholar] [CrossRef]
- Chen, L.; Wang, X.; Lu, W.; Wu, X.; Li, J. Molecular imprinting: Perspectives and applications. Chem. Soc. Rev. 2016, 45, 2137–2211. [Google Scholar] [CrossRef]
- Crespy, D.; Landfester, K. Synthesis of polyvinylpyrrolidone silver nanoparticles hybrid latex in non-aqueous miniemulsion at high temperature. Polymer 2009, 50, 1616–1620. [Google Scholar] [CrossRef]
- Crespy, D.; Landfester, K. Making dry fertile: A practical tour of non-aqueous emulsions and miniemulsions, their preparation and some applications. Soft Matter 2011, 7, 11054–11064. [Google Scholar] [CrossRef]
- Kim, H.; Kaczmarski, K.; Guiochon, G. Mass transfer kinetics on the heterogeneous binding sites of molecularly imprinted polymers. Chem. Eng. Sci. 2005, 60, 5425–5444. [Google Scholar] [CrossRef]
- Coutu, S.; Wyrsch, V.; Wynn, H.K.; Rossi, L.; Barry, D.A. Temporal dynamics of antibiotics in wastewater treatment plant influent. Sci. Total Environ. 2013, 458–460, 20–26. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Cheng, W.; Xu, L.; Strong, P.J.; Chen, H. Antibiotic-resistant genes and antibiotic-resistant bacteria in the effluent of urban residential areas, hospitals, and municipal wastewater treatment plant system. Environ. Sci. Pollut. Res. 2015, 22, 4587–4596. [Google Scholar] [CrossRef]
- Zhang, T.; Lui, F.; Chen, W.; Wang, J.; Li, K. Influence of intramolecular hydrogen bond of templates on molecular recognition of molecularly imprinted polymers. Anal. Chim. Acta 2001, 450, 53–61. [Google Scholar] [CrossRef]
- Andersson, L.I. Molecular imprinting: Developments and application in the analytical chemistry field. J. Chromatogr. B Biomed. Sci. Appl. 2000, 745, 3–13. [Google Scholar] [CrossRef]
- Soovali, L.; Room, E.; Kutt, A.; Kaljurand, I.; Leito, I. Uncertainty sources in UV-Vis spectrophotometric measurement. Accredit. Qual. Assur. 2006, 11, 246–255. [Google Scholar] [CrossRef]












| MIP/NIP | Methodology | Monomer | Crosslinker | Initiator | Solvent | Template |
|---|---|---|---|---|---|---|
| MIP 201 | Bulk | AA | TRIM | AIBN | DMSO | Amoxicillin |
| NIP 201 | Bulk | AA | TRIM | AIBN | DMSO | - |
| MIP 202 | Bulk | MAA | TRIM | AIBN | DMSO | Amoxicillin |
| NIP 202 | Bulk | MAA | TRIM | AIBN | DMSO | - |
| MIP 203 | Bulk | MAA | EGDMA | AIBN | DMSO | Amoxicillin |
| NIP 203 | Bulk | MAA | EGDMA | AIBN | DMSO | - |
| MIP 204 | Bulk | AA | EGDMA | AIBN | DMSO | Amoxicillin |
| NIP 204 | Bulk | AA | EGDMA | AIBN | DMSO | - |
| MIP 205 | Emulsion | MAA | EGDMA | AIBN | DMSO/H2O | Amoxicillin |
| NIP 205 | Emulsion | MAA | EGDMA | AIBN | DMSO/H2O | - |
| MIP/NIP | Sb/μmol g−1 (at Cf = 0.1 mM) | IF (at Cf = 0.1 mM) | Sb/μmol g−1 (at Cf = 0.2 mM) | IF (at Cf = 0.2 mM) | Sb/μmol g−1 (at Cf = 0.3 mM) | IF (at Cf = 0.3 mM) |
|---|---|---|---|---|---|---|
| 201 | MIP 5.07 NIP 2.82 | 1.80 | MIP 7.29 NIP 5.38 | 1.35 | MIP 8.80 NIP 6.41 | 1.37 |
| 202 | MIP 4.33 NIP 2.65 | 1.63 | MIP 9.74 NIP 5.00 | 1.95 | MIP 11.36 NIP 5.62 | 2.02 |
| 203 | MIP 6.36 NIP 2.49 | 2.55 | MIP 8.11 NIP 3.27 | 2.48 | MIP 8.93 NIP 3.83 | 2.33 |
| 204 | N/A | N/A | N/A | N/A | N/A | N/A |
| MIP | Compound | Sb/μmol g−1 (at Cf = 0.1 mM) | IF (at Cf = 0.1 mM) |
|---|---|---|---|
| 203 | Amoxicillin | MIP 6.36 NIP 2.49 | 2.55 |
| 205 | Amoxicillin | MIP 21.45 NIP 0.47 | 45.64 |
| 205 | Ampicillin | MIP 0.39 NIP 0.28 | 1.39 |
| 205 | Cloxacillin | MIP 8.58 NIP 7.80 | 1.1 |
| Compound | Sb/μmol g−1 (at Cf = 0.1 mM) | IF (at Cf = 0.1 mM) |
|---|---|---|
| Malachite green | MIP 19.09 NIP 15.22 | 1.25 |
| Crystal violet | MIP 36.92 NIP - | - |
| Mordant orange | MIP 7.31 NIP 3.77 | 1.94 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lowdon, J.W.; Diliën, H.; van Grinsven, B.; Eersels, K.; Cleij, T.J. Colorimetric Sensing of Amoxicillin Facilitated by Molecularly Imprinted Polymers. Polymers 2021, 13, 2221. https://0-doi-org.brum.beds.ac.uk/10.3390/polym13132221
Lowdon JW, Diliën H, van Grinsven B, Eersels K, Cleij TJ. Colorimetric Sensing of Amoxicillin Facilitated by Molecularly Imprinted Polymers. Polymers. 2021; 13(13):2221. https://0-doi-org.brum.beds.ac.uk/10.3390/polym13132221
Chicago/Turabian StyleLowdon, Joseph W, Hanne Diliën, Bart van Grinsven, Kasper Eersels, and Thomas J. Cleij. 2021. "Colorimetric Sensing of Amoxicillin Facilitated by Molecularly Imprinted Polymers" Polymers 13, no. 13: 2221. https://0-doi-org.brum.beds.ac.uk/10.3390/polym13132221