Dynamics of PHA-Accumulating Bacterial Communities Fed with Lipid-Rich Liquid Effluents from Fish-Canning Industries
Abstract
:1. Introduction
2. Materials and Methods
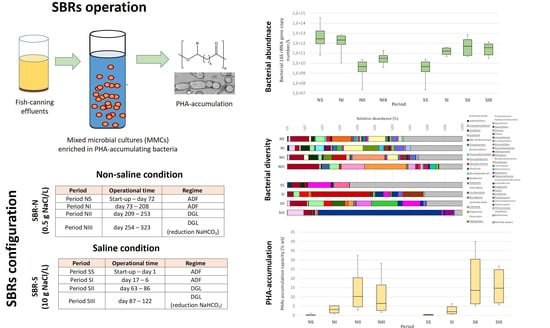
2.1. SBRs Set-Up and Operation for the PHA-Enrichment Strategy
2.2. Chemical Determinations
2.3. Microbial Analysis
2.4. Statistical Analysis
3. Results and Discussion
3.1. PHAs Accumulation Ability
3.2. Quantification of Total Bacterial Populations and Key Functional Groups
3.3. Bacterial Communities’ Diversity
3.4. Dynamics of the Bacterial Communities’ Structure
3.5. Potential PHAs Accumulation Capacities of the Dominant Bacterial Genera
3.6. Network Correlations within the Dominant Bacterial Communities
3.7. Influence of the Bacterial Communities on the PHAs Accumulation Yield
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Maheshwari, N.; Kumar, M.; Thakur, I.S.; Srivastava, S. Production, process optimization and molecular characterization of polyhydroxyalkanoate (PHA) by CO2 sequestering B. cereus SS105. Bioresour. Technol. 2018, 254, 75–82. [Google Scholar] [CrossRef] [PubMed]
- EPA. Inventory of U.S. Greenhouse Gas Emissions and Sinks 1990–2015; US Environmental Protection Agency: Washington, DC, USA, 2017.
- Sinan, M. Bioplastics for Sustainable Development: General Scenario in India. Curr. World Environ. 2020, 15, 24–28. [Google Scholar] [CrossRef]
- Naser, A.Z.; Deiab, I.; Darras, B.M. Poly(lactic acid) (PLA) and polyhydroxyalkanoates (PHAs), green alternatives to petroleum-based plastics: A review. RSC Adv. 2021, 11, 17151–17196. [Google Scholar] [CrossRef]
- Liu, C.; Luan, P.; Li, Q.; Cheng, Z.; Sun, X.; Cao, D.; Zhu, H. Biodegradable, Hygienic, and Compostable Tableware from Hybrid Sugarcane and Bamboo Fibers as Plastic Alternative. Matter 2020, 3, 2066–2079. [Google Scholar] [CrossRef]
- Shah, S.; Matkawala, F.; Garg, S.; Nighojkar, S.; Nighojkar, A.; Kumar, A. Emerging Trend of Bio-plastics and Its Impact on Society. Biotechnol. J. Int. 2020, 24, 1–10. [Google Scholar] [CrossRef]
- Tsang, Y.F.; Kumar, V.; Samadar, P.; Yang, Y.; Lee, J.; Ok, Y.S.; Song, H.; Kim, K.-H.; Kwon, E.E.; Jeon, Y.J. Production of bioplastic through food waste valorization. Environ. Int. 2019, 127, 625–644. [Google Scholar] [CrossRef]
- Khoo, H.H.; Tan, R.B.H.; Chng, K.W.L. Environmental impacts of conventional plastic and bio-based carrier bags. Int. J. Life Cycle Assess. 2010, 15, 284–293. [Google Scholar] [CrossRef]
- Choi, S.Y.; Cho, I.J.; Lee, Y.; Kim, Y.; Kim, K.; Lee, S.Y. Microbial Polyhydroxyalkanoates and Nonnatural Polyesters. Adv. Mater. 2020, 32, 1907138. [Google Scholar] [CrossRef]
- Grujić, R.; Vujadinović, D.; Savanović, D. Biopolymers as food packaging materials. In Advances in Applications of Industrial Biomaterials; Pellicer, E., Nikolic, D., Sort, J., Baró, M., Zivic, F., Grujovic, N., Grujic, R., Pelemis, S., Eds.; Springer: Cham, Switzerland, 2017; pp. 139–160. [Google Scholar]
- Tugarova, A.V.; Dyatlova, Y.A.; Kenzhegulov, O.A.; Kamnev, A.A. Poly-3-hydroxybutyrate synthesis by different Azospirillum brasilense strains under varying nitrogen deficiency: A comparative in-situ FTIR spectroscopic analysis. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2021, 252, 119458. [Google Scholar] [CrossRef]
- Tsuge, T.; Hyakutake, M.; Mizuno, K. Class IV polyhydroxyalkanoate (PHA) synthases and PHA-producing Bacillus. Appl. Microbiol. Biotechnol. 2015, 99, 6231–6240. [Google Scholar] [CrossRef]
- Kaewbai-Ngam, A.; Incharoensakdi, A.; Monshupanee, T. Increased accumulation of polyhydroxybutyrate in divergent cyanobacteria under nutrient-deprived photoautotrophy: An efficient conversion of solar energy and carbon dioxide to polyhydroxybutyrate by Calothrix scytonemicola TISTR 8095. Bioresour. Technol. 2016, 212, 342–347. [Google Scholar] [CrossRef] [PubMed]
- Sheu, D.-S.; Lai, Y.-W.; Chang, R.-C.; Chen, W.-M. Detection of polyhydroxyalkanoate synthase activity on a polyacrylamide gel. Anal. Biochem. 2009, 393, 62–66. [Google Scholar] [CrossRef] [PubMed]
- Reddy, M.V.; Mawatari, Y.; Onodera, R.; Nakamura, Y.; Yajima, Y.; Chang, Y.-C. Polyhydroxyalkanoates (PHA) production from synthetic waste using Pseudomonas pseudoflava: PHA synthase enzyme activity analysis from P. pseudoflava and P. palleronii. Bioresour. Technol. 2017, 234, 99–105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Govil, T.; Wang, J.; Samanta, D.; David, A.; Tripathi, A.; Rauniyar, S.; Salem, D.R.; Sani, R.K. Lignocellulosic feedstock: A review of a sustainable platform for cleaner production of nature’s plastics. J. Clean. Prod. 2020, 270, 122521. [Google Scholar] [CrossRef]
- Huang, L.; Chen, Z.; Wen, Q.; Zhao, L.; Lee, D.-J.; Yang, L.; Wang, Y. Insights into Feast-Famine polyhydroxyalkanoate (PHA)-producer selection: Microbial community succession, relationships with system function and underlying driving forces. Water Res. 2018, 131, 167–176. [Google Scholar] [CrossRef]
- Villano, M.; Beccari, M.; Dionisi, D.; Lampis, S.; Miccheli, A.; Vallini, G.; Majone, M. Effect of pH on the production of bacterial polyhydroxyalkanoates by mixed cultures enriched under periodic feeding. Process Biochem. 2010, 45, 714–723. [Google Scholar] [CrossRef]
- Kourmentza, C.; Plácido, J.; Venetsaneas, N.; Burniol-Figols, A.; Varrone, C.; Gavala, H.N.; Reis, M.A.M. Recent Advances and Challenges towards Sustainable Polyhydroxyalkanoate (PHA) Production. Bioengineering 2017, 4, 55. [Google Scholar] [CrossRef] [Green Version]
- Queirós, D.; Rangel, C.; Lemos, P.C.; Rossetti, S.; Serafim, L.S. Impact of Organic Acids Supplementation to Hardwood Spent Sulfite Liquor as Substrate for the Selection of Polyhydroxyalkanoates-Producing Organisms. Fermentation 2018, 4, 58. [Google Scholar] [CrossRef] [Green Version]
- Patel, M.; Gapes, D.J.; Newman, R.H.; Dare, P.H. Physico-chemical properties of polyhydroxyalkanoate produced by mixed-culture nitrogen-fixing bacteria. Appl. Microbiol. Biotechnol. 2009, 82, 545–555. [Google Scholar] [CrossRef]
- Chen, Z.; Guo, Z.; Wen, Q.; Huang, L.; Bakke, R.; Du, M. A new method for polyhydroxyalkanoate (PHA) accumulating bacteria selection under physical selective pressure. Int. J. Biol. Macromol. 2015, 72, 1329–1334. [Google Scholar] [CrossRef]
- De Souza Reis, G.A.; Michels, M.H.A.; Fajardo, G.L.; Lamot, I.; De Best, J.H. Optimization of Green Extraction and Purification of PHA Produced by Mixed Microbial Cultures from Sludge. Water 2020, 12, 1185. [Google Scholar] [CrossRef] [Green Version]
- Rogala, M.M.; Gawor, J.; Gromadka, R.; Kowalczyk, M.; Grzesiak, J. Biodiversity and Habitats of Polar Region Polyhydroxyalkanoic Acid-Producing Bacteria: Bioprospection by Popular Screening Methods. Genes 2020, 11, 873. [Google Scholar] [CrossRef] [PubMed]
- Sruamsiri, D.; Thayanukul, P.; Suwannasilp, B.B. In situ identification of polyhydroxyalkanoate (PHA)-accumulating microorganisms in mixed microbial cultures under feast/famine conditions. Sci. Rep. 2020, 10, 3752. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Del Oso, M.S.; Mauricio-Iglesias, M.; Hospido, A. Evaluation and optimization of the environmental performance of PHA downstream processing. Chem. Eng. J. 2021, 412, 127687. [Google Scholar] [CrossRef]
- Tamis, J.; Mulders, M.; Dijkman, H.; Rozendal, R.; Van Loosdrecht, M.C.M.; Kleerebezem, R. Pilot-Scale Polyhydroxyalkanoate Production from Paper Mill Wastewater: Process Characteristics and Identification of Bottlenecks for Full-Scale Implementation. J. Environ. Eng. 2018, 144, 04018107. [Google Scholar] [CrossRef]
- Wang, X.; Oehmen, A.; Carvalho, G.; Reis, M.A. Community profile governs substrate competition in polyhydroxyalkanoate (PHA)-producing mixed cultures. New Biotechnol. 2020, 58, 32–37. [Google Scholar] [CrossRef]
- Saratale, R.G.; Cho, S.-K.; Saratale, G.D.; Kadam, A.A.; Ghodake, G.S.; Kumar, M.; Bharagava, R.N.; Kumar, G.; Kim, D.S.; Mulla, S.I.; et al. A comprehensive overview and recent advances on polyhydroxyalkanoates (PHA) production using various organic waste streams. Bioresour. Technol. 2021, 325, 124685. [Google Scholar] [CrossRef]
- Sen, K.Y.; Baidurah, S. Renewable biomass feedstocks for production of sustainable biodegradable polymer. Curr. Opin. Green Sustain. Chem. 2021, 27, 100412. [Google Scholar] [CrossRef]
- Stahel, W.R. The circular economy. Nature 2016, 531, 435–438. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez-Perez, S.; Serrano, A.; Pantión, A.A.; Alonso-Fariñas, B. Challenges of scaling-up PHA production from waste streams. A review. J. Environ. Manag. 2018, 205, 215–230. [Google Scholar] [CrossRef] [Green Version]
- Horikoshi, K.; Antranikian, G.; Bull, A.T.; Robb, F.T.; Stetter, K.O. Extremophiles Handbook; Horikoshi, K., Ed.; Springer: Tokyo, Japan, 2010. [Google Scholar]
- Correa-Galeote, D.; Roibás, A.; Mosquera-Corral, A.; Juárez-Jiménez, B.; González-López, J.; Rodelas, B. Salinity is the major driver of the global eukaryotic community structure in fish-canning wastewater treatment plants. J. Environ. Manag. 2021, 290, 112623. [Google Scholar] [CrossRef] [PubMed]
- Correa-Galeote, D.; Roibás-Rozas, A.; Mosquera-Corral, A.; Juárez-Jiménez, B.; González-López, J.; Rodelas, B. Revealing the dissimilar structure of microbial communities in different WWTPs that treat fish-canning wastewater with different NaCl content. J. Water Process Eng. 2021, 44, 102328. [Google Scholar] [CrossRef]
- Argiz, L.; Gonzalez-Cabaleiro, R.; Correa-Galeote, D.; del Rio, A.V.; Mosquera-Corral, A. Open-culture biotechnological process for triacylglycerides and polyhydroxyalkanoates recovery from industrial waste fish oil under saline conditions. Sep. Purif. Technol. 2021, 270, 118805. [Google Scholar] [CrossRef]
- Argiz, L.; González-Cabaleiro, R.; del Río, Á.V.; González-López, J.; Mosquera-Corral, A. A novel strategy for triacylglycerides and polyhydroxyalkanoates production using waste lipids. Sci. Total Environ. 2021, 763, 142944. [Google Scholar] [CrossRef] [PubMed]
- Wen, Q.; Liu, S.; Liu, Y.; Chen, Z. Effect of inoculum and organic loading on mixed culture polyhydroxyalkanoate production using crude glycerol as the substrate. Int. J. Biol. Macromol. 2021, 182, 1785–1792. [Google Scholar] [CrossRef] [PubMed]
- Sangkharak, K.; Paichid, N.; Yunu, T.; Klomklao, S.; Prasertsan, P. Utilisation of tuna condensate waste from the canning industry as a novel substrate for polyhydroxyalkanoate production. Biomass Convers. Biorefin. 2021, 11, 2053–2064. [Google Scholar] [CrossRef]
- Van Thuoc, D.; My, D.N.; Loan, T.T.; Sudesh, K. Utilization of waste fish oil and glycerol as carbon sources for polyhydroxyalkanoate production by Salinivibrio sp. M318. Int. J. Biol. Macromol. 2019, 141, 885–892. [Google Scholar] [CrossRef]
- Oliveira, C.S.S.; Silva, C.E.; Carvalho, G.; Reis, M.A. Strategies for efficiently selecting PHA producing mixed microbial cultures using complex feedstocks: Feast and famine regime and uncoupled carbon and nitrogen availabilities. New Biotechnol. 2017, 37, 69–79. [Google Scholar] [CrossRef]
- Lorini, L.; di Re, F.; Majone, M.; Valentino, F. High rate selection of PHA accumulating mixed cultures in sequencing batch reactors with uncoupled carbon and nitrogen feeding. New Biotechnol. 2020, 56, 140–148. [Google Scholar] [CrossRef]
- APHA. Standard Methods for the Examination of Water and Wastewater, 21st ed.; Rice, E.W., Baird, R.B., Eaton, A.D., Clesceri, L.S., Eds.; American Public Health Association: Washington, DC, USA, 2005. [Google Scholar]
- Takahashi, S.; Tomita, J.; Nishioka, K.; Hisada, T.; Nishijima, M. Development of a Prokaryotic Universal Primer for Simultaneous Analysis of Bacteria and Archaea Using Next-Generation Sequencing. PLoS ONE 2014, 9, e105592. [Google Scholar] [CrossRef] [Green Version]
- Katoh, K.; Asimenos, G.; Toh, H. Multiple alignment of DNA sequences with MAFFT. In Bioinformatics for DNA Sequence Analysis; Posada, D., Ed.; Springer: New York, NY, USA, 2009; pp. 39–64. [Google Scholar]
- Hill, T.C.; Walsh, K.A.; Harris, J.A.; Moffett, B.F. Using ecological diversity measures with bacterial communities. FEMS Microbiol. Ecol. 2003, 43, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Novelli, L.D.D.; Sayavedra, S.M.; Rene, E.R. Polyhydroxyalkanoate (PHA) production via resource recovery from industrial waste streams: A review of techniques and perspectives. Bioresour. Technol. 2021, 331, 124985. [Google Scholar] [CrossRef] [PubMed]
- De Cocker, P.; Bessiere, Y.; Hernandez-Raquet, G.; Dubos, S.; Mozo, I.; Gaval, G.; Caligaris, M.; Barillon, B.; Vlaeminck, S.; Sperandio, M. Enrichment and adaptation yield high anammox conversion rates under low temperatures. Bioresour. Technol. 2018, 250, 505–512. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.; Leary, E.O.; Behrens, S.; LaPara, T.M. Evaluating Quantitative PCR Assays to Enumerate Several Bacterial Populations of Importance in Different Municipal Wastewater Treatment Designs. J. Environ. Eng. 2021, 147, 04021044. [Google Scholar] [CrossRef]
- Jurburg, S.D.; Nunes, I.; Stegen, J.C.; LE Roux, X.; Priemé, A.; Sørensen, S.; Salles, J.F. Autogenic succession and deterministic recovery following disturbance in soil bacterial communities. Sci. Rep. 2017, 7, 45691. [Google Scholar] [CrossRef] [Green Version]
- De los Reyes, F.L., III; Raskin, L. Role of filamentous microorganisms in activated sludge foaming: Relationship of mycolata levels to foaming initiation and stability. Water Res. 2002, 36, 445–459. [Google Scholar] [CrossRef]
- Oshiki, M.; Satoh, H.; Mino, T.; Onuki, M. PHA-accumulating microorganisms in full-scale wastewater treatment plants. Water Sci. Technol. 2008, 58, 13–20. [Google Scholar] [CrossRef] [Green Version]
- Marzorati, M.; Wittebolle, L.; Boon, N.; Daffonchio, D.; Verstraete, W. How to get more out of molecular fingerprints: Practical tools for microbial ecology. Environ. Microbiol. 2008, 10, 1571–1581. [Google Scholar] [CrossRef]
- Pereira, J.; Queirós, D.; Lemos, P.C.; Rossetti, S.; Serafim, L.S. Enrichment of a mixed microbial culture of PHA-storing microorganisms by using fermented hardwood spent sulfite liquor. New Biotechnol. 2020, 56, 79–86. [Google Scholar] [CrossRef]
- Morgan-Sagastume, F.; Hjort, M.; Cirne, D.; Gérardin, F.; Lacroix, S.; Gaval, G.; Karabegovic, L.; Alexandersson, T.; Johansson, P.; Karlsson, A.; et al. Integrated production of polyhydroxyalkanoates (PHAs) with municipal wastewater and sludge treatment at pilot scale. Bioresour. Technol. 2015, 181, 78–89. [Google Scholar] [CrossRef]
- Albuquerque, M.G.E.; Eiroa, M.; Torres, C.; Nunes, B.R.; Reis, M.A.M. Strategies for the development of a side stream process for polyhydroxyalkanoate (PHA) production from sugar cane molasses. J. Biotechnol. 2007, 130, 411–421. [Google Scholar] [CrossRef]
- Alvarenga, D.; Andreote, A.P.D.; Branco, L.H.; Fiore, M.F. Kryptousia macronema gen. nov., sp. nov. and Kryptousia microlepis sp. nov., nostocalean cyanobacteria isolated from phyllospheres. Int. J. Syst. Evol. Microbiol. 2017, 67, 3301–3309. [Google Scholar] [CrossRef] [Green Version]
- Bazylinski, D.A.; Blakemore, R.P. Nitrogen fixation (acetylene reduction) inAquaspirillum magnetotacticum. Curr. Microbiol. 1983, 9, 305–308. [Google Scholar] [CrossRef]
- Krotzky, A.; Werner, D. Nitrogen fixation in Pseudomonas stutzeri. Arch. Microbiol. 1987, 147, 48–57. [Google Scholar] [CrossRef]
- Pankievicz, V.C.S.; Amaral, F.P.D.; Ane, J.-M.; Stacey, G. Diazotrophic Bacteria and Their Mechanisms to Interact and Benefit Cereals. Mol. Plant-Microbe Interact. 2021, 34, 491–498. [Google Scholar] [CrossRef]
- Ramos, P.L.; Van Trappen, S.; Thompson, F.L.; Rocha, R.C.S.; Barbosa, H.R.; De Vos, P.; Moreira-Filho, C.A. Screening for endophytic nitrogen-fixing bacteria in Brazilian sugar cane varieties used in organic farming and description of Stenotrophomonas pavanii sp. nov. Int. J. Syst. Evol. Microbiol. 2011, 61, 926–931. [Google Scholar] [CrossRef] [Green Version]
- Sachdev, D.; Nema, P.; Dhakephalkar, P.; Zinjarde, S.; Chopade, B. Assessment of 16S rRNA gene-based phylogenetic diversity and promising plant growth-promoting traits of Acinetobacter community from the rhizosphere of wheat. Microbiol. Res. 2010, 165, 627–638. [Google Scholar] [CrossRef]
- Smit, A.-M.; Strabala, T.J.; Peng, L.; Rawson, P.; Lloyd-Jones, G.; Jordan, T.W. Proteomic Phenotyping of Novosphingobium nitrogenifigens Reveals a Robust Capacity for Simultaneous Nitrogen Fixation, Polyhydroxyalkanoate Production, and Resistance to Reactive Oxygen Species. Appl. Environ. Microbiol. 2012, 78, 4802–4815. [Google Scholar] [CrossRef] [Green Version]
- Stewart, W.D.P. Nitrogen Fixation by Myxophyceae from Marine Environments. Microbiology 1964, 36, 415–422. [Google Scholar] [CrossRef] [Green Version]
- Encarnación, S.; Vargas, M.D.C.; Dunn, M.F.; Dávalos, A.; Mendoza, G.; Mora, Y.; Mora, J. AniA Regulates Reserve Polymer Accumulation and Global Protein Expression in Rhizobium etli. J. Bacteriol. 2002, 184, 2287–2295. [Google Scholar] [CrossRef] [Green Version]
- Bhati, R.; Mallick, N. Production and characterization of poly(3-hydroxybutyrate-co-3-hydroxyvalerate) co-polymer by a N2-fixing cyanobacterium, Nostoc muscorum Agardh. J. Chem. Technol. Biotechnol. 2012, 87, 505–512. [Google Scholar] [CrossRef]
- Okon, Y.; Itzigsohn, R. Poly-β-hydroxybutyrate metabolism in Azospirillum brasilense and the ecological role of PHB in the rhizosphere. FEMS Microbiol. Rev. 1992, 9, 131–139. [Google Scholar] [CrossRef]
- Obruca, S.; Sedlacek, P.; Slaninova, E.; Fritz, I.; Daffert, C.; Meixner, K.; Sedrlova, Z.; Koller, M. Novel unexpected functions of PHA granules. Appl. Microbiol. Biotechnol. 2020, 104, 4795–4810. [Google Scholar] [CrossRef]
- Skorupski, M.; Butkiewicz, G.; Wierzbicka, A. The first reaction of soil mite fauna (Acari, Mesostigmata) caused by conversion of Norway spruce stand in the Szklarska Poręba Forest District. J. For. Sci. 2009, 55, 235–243. [Google Scholar] [CrossRef] [Green Version]
- Lucena-Padrós, H.; Ruiz-Barba, J.L. Microbial biogeography of Spanish-style green olive fermentations in the province of Seville, Spain. Food Microbiol. 2019, 82, 259–268. [Google Scholar] [CrossRef]
- Allison, S.D.; Martiny, J.B.H. Resistance, resilience, and redundancy in microbial communities. Proc. Natl. Acad. Sci. USA 2008, 105, 11512–11519. [Google Scholar] [CrossRef] [Green Version]
- Schulze, R.; Spring, S.; Amann, R.; Huber, I.; Ludwig, W.; Schleifer, K.-H.; Kampfer, P. Genotypic Diversity of Acidovorax Strains Isolated from Activated Sludge and Description of Acidovorax defluvii sp. nov. Syst. Appl. Microbiol. 1999, 22, 205–214. [Google Scholar] [CrossRef]
- Anburajan, P.; Kumar, A.N.; Sabapathy, P.C.; Kim, G.-B.; Cayetano, R.D.; Yoon, J.-J.; Kumar, G.; Kim, S.-H. Polyhydroxy butyrate production by Acinetobacter junii BP25, Aeromonas hydrophila ATCC 7966, and their co-culture using a feast and famine strategy. Bioresour. Technol. 2019, 293, 122062. [Google Scholar] [CrossRef]
- Pramanik, N.; Mukherjee, K.; Nandy, A.; Mukherjee, S.; Kundu, P.P. Comparative analysis of different properties of polyhydroxyalkanoates isolated from two different bacterial strains: Alkaliphilus oremlandii OhILAs and recombinant Escherichia coli XL1B. J. Appl. Polym. Sci. 2014, 131, 41080. [Google Scholar] [CrossRef]
- Mohammadi, M.; Hassan, M.A.; Shirai, Y.; Man, H.C.; Ariffin, H.; Yee, L.-N.; Mumtaz, T.; Chong, M.-L.; Phang, L.-Y. Separation and Purification of Polyhydroxyalkanoates from Newly Isolated Comamonas sp. EB172 by Simple Digestion with Sodium Hydroxide. Sep. Sci. Technol. 2012, 47, 534–541. [Google Scholar] [CrossRef]
- Anderson, A.J.; Haywood, G.W.; Williams, D.R.; Dawes, E.A. The production of polyhydroxyalkanoates from unrelated car-bon sources. In Novel Biodegradable Microbial Polymers; Dawes, E.A., Ed.; Kluwer Academic Publishers: Dordrecht, The Netherlands, 1990; pp. 119–129. [Google Scholar]
- Sheu, S.-Y.; Sheu, D.-S.; Sheu, F.-S.; Chen, W.-M. Gemmobacter tilapiae sp. nov., a poly-β-hydroxybutyrate-accumulating bacterium isolated from a freshwater pond. Int. J. Syst. Evol. Microbiol. 2013, 63, 1550–1556. [Google Scholar] [CrossRef] [Green Version]
- Kumar, V.; Thakur, V.; Ambika; Kumar, S.; Singh, D. Bioplastic reservoir of diverse bacterial communities revealed along altitude gradient of Pangi-Chamba trans-Himalayan region. FEMS Microbiol. Lett. 2018, 365, fny144. [Google Scholar] [CrossRef] [Green Version]
- Gholami, A.; Ghasemi, Y.; Kazemi, A.; Abootalebi, S.N.; Irajie, C.; Ireji, A.; Omidifar, N.; Mohkam, M. Bacterial Strain Isolated from High-Salt Environments Can Produce Large Amounts of New Polyhydroxyalkanoate (PHA). J. Environ. Treat. Tech. 2020, 8, 1268–1273. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, Y.; Lin, Y.; Wang, L.; Yao, S.; Cao, Y.; Zhai, L.; Tang, X.; Zhang, L.; Zhang, T.; et al. Novosphingobium clariflavum sp. nov., isolated from a household product plant. Int. J. Syst. Evol. Microbiol. 2017, 67, 3150–3155. [Google Scholar] [CrossRef]
- Gaisin, V.A.; Burganskaya, E.I.; Grouzdev, D.S.; Osipova, N.S.; Ashikhmin, A.A.; Sinetova, M.A.; Krutkina, M.S.; Bryantseva, I.A.; Sukhacheva, M.V.; Kochetkova, T.V.; et al. ‘Candidatus Oscillochloris fontis’: A novel mesophilic phototrophic Chloroflexota bacterium belonging to the ubiquitous Oscillochloris genus. FEMS Microbiol. Lett. 2019, 366, fnz097. [Google Scholar] [CrossRef]
- De Paula, F.C.; Kakazu, S.; de Paula, C.B.C.; Gomez, J.G.C.; Contiero, J. Polyhydroxyalkanoate production from crude glycerol by newly isolated Pandoraea sp. J. King Saud Univ.-Sci. 2017, 29, 166–173. [Google Scholar] [CrossRef]
- Srivastava, A.; Murugaiyan, J.; Garcia, J.A.L.; De Corte, D.; Hoetzinger, M.; Eravci, M.; Weise, C.; Kumar, Y.; Roesler, U.; Hahn, M.W.; et al. Combined Methylome, Transcriptome and Proteome Analyses Document Rapid Acclimatization of a Bacterium to Environmental Changes. Front. Microbiol. 2020, 11, 2197. [Google Scholar] [CrossRef]
- Yao, J.; Zhang, G.; Wu, Q.; Chen, G.-Q.; Zhang, R. Production of polyhydroxyalkanoates by Pseudomonas nitroreducens. Antonie Leeuwenhoek 1999, 75, 345–349. [Google Scholar] [CrossRef]
- Kosmachevskaya, O.V.; Osipov, E.V.; Van Chi, T.; Mai, P.T.T.; Topunov, A.F. Effect of Cultivation Conditions on Poly(3-hydroxybutyrate) Synthesis by Nodule Bacteria Rhizobium phaseoli. Appl. Biochem. Microbiol. 2020, 56, 64–71. [Google Scholar] [CrossRef]
- Kim, Y.-M.; Nowack, S.; Olsen, M.T.; Becraft, E.D.; Wood, J.; Ethiel, V.; Eklapper, I.; Kühl, M.; Fredrickson, J.K.; Bryant, D.A.; et al. Diel metabolomics analysis of a hot spring chlorophototrophic microbial mat leads to new hypotheses of community member metabolisms. Front. Microbiol. 2015, 6, 209. [Google Scholar] [CrossRef]
- Clifton-García, B.; González-Reynoso, O.; Robledo-Ortíz, J.R.; Villafaña-Rojas, J.; González-García, Y. Forest soil bacteria able to produce homo and copolymers of polyhydroxyalkanoates from several pure and waste carbon sources. Lett. Appl. Microbiol. 2020, 70, 300–309. [Google Scholar] [CrossRef] [PubMed]
- Khammas, K.; Ageron, E.; Grimont, P.; Kaiser, P. Azospirillum irakense sp. nov., a nitrogen-fixing bacterium associated with rice roots and rhizosphere soil. Res. Microbiol. 1989, 140, 679–693. [Google Scholar] [CrossRef]
- Mezzolla, V.; D’Urso, O.F.; Poltronieri, P. Role of PhaC Type I and Type II Enzymes during PHA Biosynthesis. Polymers 2018, 10, 910. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, D.; Li, J.; Ma, X. Accumulation of bioplastic polyhydroxyalkanoate with different substrate forms from pretreated waste lignocellulose hydrolysate. Ind. Crop. Prod. 2021, 172, 114061. [Google Scholar] [CrossRef]
- Woraittinun, N.; Suwannasilp, B.B. polyhydroxyalkanoate production from different carbon substrates using sludge from a wastewater treatment plant: Microbial communities, polymer compositions, and thermal characteristics. Environ. Prog. Sustain. Energy 2017, 36, 1754–1764. [Google Scholar] [CrossRef]
- Yan, X.; Li, D.; Ma, X.; Li, J. Bioconversion of renewable lignocellulosic biomass into multicomponent substrate via pressurized hot water pretreatment for bioplastic polyhydroxyalkanoate accumulation. Bioresour. Technol. 2021, 339, 125667. [Google Scholar] [CrossRef]
- Yin, F.; Li, D.; Ma, X.; Li, J.; Qiu, Y. Poly(3-hydroxybutyrate-3-hydroxyvalerate) production from pretreated waste lignocellulosic hydrolysates and acetate co-substrate. Bioresour. Technol. 2020, 316, 123911. [Google Scholar] [CrossRef]
- Martínez, V.; de la Peña, F.; García-Hidalgo, J.; de la Mata, I.; García, J.L.; Prieto, M.A. Identification and Biochemical Evidence of a Medium-Chain-Length Polyhydroxyalkanoate Depolymerase in the Bdellovibrio bacteriovorus Predatory Hydrolytic Arsenal. Appl. Environ. Microbiol. 2012, 78, 6017–6026. [Google Scholar] [CrossRef] [Green Version]
- Martínez, V.; Herencias, C.; Jurkevitch, E.; Prieto, M.A. Engineering a predatory bacterium as a proficient killer agent for intracellular bio-products recovery: The case of the polyhydroxyalkanoates. Sci. Rep. 2016, 6, 24381. [Google Scholar] [CrossRef] [Green Version]
- Gahlawat, G. (Ed.) Polyhydroxyalkanoates: The future bioplastics. In Polyhydroxyalkanoates Biopolymers; Springer: Cham, Switzerland, 2019; pp. 15–23. [Google Scholar]
- Feng, X.-M.; Mo, Y.-X.; Han, L.; Nogi, Y.; Zhu, Y.-H.; Lv, J. Qipengyuania sediminis gen. nov., sp. nov., a member of the family Erythrobacteraceae isolated from subterrestrial sediment. Int. J. Syst. Evol. Microbiol. 2015, 65, 3658–3665. [Google Scholar] [CrossRef]
- Favaro, L.; Basaglia, M.; Rodriguez, J.E.G.; Morelli, A.; Ibraheem, O.; Pizzocchero, V.; Casella, S. Bacterial production of PHAs from lipid-rich by-products. Appl. Food Biotechnol. 2019, 6, 45–52. [Google Scholar] [CrossRef]
- Heylen, K.; Lebbe, L.; De Vos, P. Acidovorax caeni sp. nov., a denitrifying species with genetically diverse isolates from activated sludge. Int. J. Syst. Evol. Microbiol. 2008, 58, 73–77. [Google Scholar] [CrossRef]
- Snellman, E.A.; Sullivan, E.R.; Colwell, R.R. Purification and properties of the extracellular lipase, LipA, of Acinetobacter sp. RAG-1. Eur. J. Biochem. 2002, 269, 5771–5779. [Google Scholar] [CrossRef] [PubMed]
- Haiyambo, D.H.; Chimwamurombe, P.M. Isolation of the Azospirillum Species from the Rhizosphere of the Leguminous Bauhinia petersiana in North Eastern Namibia. Jordan J. Biol. Sci. 2018, 11, 347–353. [Google Scholar]
- Pal, M.; Swarnkar, M.K.; Dhar, H.; Chhibber, S.; Gulati, A. Genome assembly of Chryseobacterium sp. strain IHBB 10212 from glacier top-surface soil in the Indian trans-Himalayas with potential for hydrolytic enzymes. Genom. Data 2017, 13, 46–49. [Google Scholar] [CrossRef] [PubMed]
- Chow, W.S.; Tan, S.G.; Ahmad, Z.A.; Chia, K.H.; Lau, N.S.; Sudesh, K. Biodegradability of Epoxidized Soybean Oil Based Thermosets in Compost Soil Environment. J. Polym. Environ. 2014, 22, 140–147. [Google Scholar] [CrossRef]
- Schröder, J.; Glaub, A.; Schneider, J.; Trost, E.; Tauch, A. Draft Genome Sequence of Corynebacterium bovis DSM 20582, Which Causes Clinical Mastitis in Dairy Cows. J. Bacteriol. 2012, 194, 4437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.D. Devosia subaequoris sp. nov., isolated from beach sediment. Int. J. Syst. Evol. Microbiol. 2007, 57, 2212–2215. [Google Scholar] [CrossRef] [Green Version]
- Yoon, J.-H.; Kang, S.-J.; Oh, T.-K. Dokdonella koreensis gen. nov., sp. nov., isolated from soil. Int. J. Syst. Evol. Microbiol. 2006, 56, 145–150. [Google Scholar] [CrossRef] [Green Version]
- Xie, C.-H.; Yokota, A. Dyella japonica gen. nov., sp. nov., a γ-proteobacterium isolated from soil. Int. J. Syst. Evol. Microbiol. 2005, 55, 753–756. [Google Scholar] [CrossRef] [Green Version]
- Kang, H.; Kim, H.; Joung, Y.; Jang, T.Y.; Joh, K. Ferruginibacter paludis sp. nov., isolated from wetland freshwater, and emended descriptions of Ferruginibacter lapsinanis and Ferruginibacter alkalilentus. Int. J. Syst. Evol. Microbiol. 2015, 65, 2635–2639. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Q.; Qian, J.; Ma, L. Mutation breeding of lipase-producing strain Flavobacterium sp. by supercritical CO2 with hydrazine hydrate. Braz. Arch. Biol. Technol. 2013, 56, 541–546. [Google Scholar] [CrossRef] [Green Version]
- Indest, K.J.; Eberly, J.; Ringelberg, D.B.; Hancock, D.E. The effects of putative lipase and wax ester synthase/acyl-CoA:diacylglycerol acyltransferase gene knockouts on triacylglycerol accumulation in Gordonia sp. KTR9. J. Ind. Microbiol. Biotechnol. 2015, 42, 219–227. [Google Scholar] [CrossRef]
- Battu, L.; Ulaganathan, K. Whole genome sequencing and identification of host-interactive genes in the rice endophytic Leifsonia sp. ku-ls. Funct. Integr. Genom. 2020, 20, 237–243. [Google Scholar] [CrossRef] [PubMed]
- Ko, H.-S.; Jin, R.-D.; Krishnan, H.B.; Lee, S.-B.; Kim, K.-Y. Biocontrol Ability of Lysobacter antibioticus HS124 Against Phytophthora Blight Is Mediated by the Production of 4-Hydroxyphenylacetic Acid and Several Lytic Enzymes. Curr. Microbiol. 2009, 59, 608–615. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.-Y.; Hameed, A.; Shen, F.-T.; Liu, Y.-C.; Hsu, Y.-H.; Shahina, M.; Lai, W.-A.; Young, C.-C. Description of Niveispirillum fermenti gen. nov., sp. nov., isolated from a fermentor in Taiwan, transfer of Azospirillum irakense (1989) as Niveispirillum irakense comb. nov., and reclassification of Azospirillum amazonense (1983) as Nitrospirillum amazonense gen. nov. Antonie Leeuwenhoek 2014, 105, 1149–1162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Pei, T.; Du, J.; Huang, H.; Deng, M.-R.; Zhu, H. Comparative genomic analysis of the genus Novosphingobium and the description of two novel species Novosphingobium aerophilum sp. nov. and Novosphingobium jiangmenense sp. nov. Syst. Appl. Microbiol. 2021, 44, 126202. [Google Scholar] [CrossRef]
- Coenye, T.; Falsen, E.; Hoste, B.; Ohlén, M.; Goris, J.; Govan, J.R.; Gillis, M.; Vandamme, P. Description of Pandoraea gen. nov. with Pandoraea apista sp. nov., Pandoraea pulmonicola sp. nov., Pandoraea pnomenusa sp. nov., Pandoraea sputorum sp. nov. and Pandoraea norimbergensis comb. nov. Int. J. Syst. Evol. Microbiol. 2000, 50, 887–899. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Pei, T.; Deng, M.-R.; Zhu, H. Qipengyuania soli sp. nov., Isolated from Mangrove Soil. Curr. Microbiol. 2021, 78, 2806–2814. [Google Scholar] [CrossRef]
- Datta, A.; Singh, R.K.; Tabassum, S. Isolation, characterization and growth of Rhizobium strains under optimum conditions for effective biofertilizer production. Int. J. Pharm. Sci. Rev. Res. 2015, 32, 199–208. [Google Scholar]
- Hasan-Beikdashti, M.; Forootanfar, H.; Safiarian, M.; Ameri, A.; Ghahremani, M.; Khoshayand, M.; Faramarzi, M. Optimization of culture conditions for production of lipase by a newly isolated bacterium Stenotrophomonas maltophilia. J. Taiwan Inst. Chem. Eng. 2012, 43, 670–677. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, Y.; Wei, L.; Wang, Y.; Shen, X.; Li, S. Taibaiella smilacinae gen. nov., sp. nov., an endophytic member of the family Chitinophagaceae isolated from the stem of Smilacina japonica, and emended description of Flavihumibacter petaseus. Int. J. Syst. Evol. Microbiol. 2013, 63, 3769–3776. [Google Scholar] [CrossRef]
- Abdollahi, P.; Ghane, M.; Babaeekhou, L. Isolation and Characterization of Thermophilic Bacteria from Gavmesh Goli Hot Spring in Sabalan Geothermal Field, Iran: Thermomonas hydrothermalis and Bacillus altitudinis Isolates as a Potential Source of Thermostable Protease. Geomicrobiol. J. 2021, 38, 87–95. [Google Scholar] [CrossRef]
- Huang, J.; Yan, R.; He, J.-Y.; Wang, P. Purification and Immobilization of a Novel Enantioselective Lipase from Tsukamurella tyrosinosolvents for Efficient Resolution of Ethyl 2-(2-oxopyrrolidin-1-yl) Butyrate. Appl. Biochem. Biotechnol. 2016, 180, 609–622. [Google Scholar] [CrossRef] [PubMed]
- Layeghifard, M.; Hwang, D.M.; Guttman, D.S. Disentangling Interactions in the Microbiome: A Network Perspective. Trends Microbiol. 2017, 25, 217–228. [Google Scholar] [CrossRef]
- Qian, X.; Li, H.; Wang, Y.; Wu, B.; Wu, M.; Chen, L.; Li, X.; Zhang, Y.; Wang, X.; Shi, M.; et al. Leaf and Root Endospheres Harbor Lower Fungal Diversity and Less Complex Fungal Co-occurrence Patterns Than Rhizosphere. Front. Microbiol. 2019, 10, 1015. [Google Scholar] [CrossRef] [PubMed]
- Crognale, S.; Tonanzi, B.; Valentino, F.; Majone, M.; Rossetti, S. Microbiome dynamics and phaC synthase genes selected in a pilot plant producing polyhydroxyalkanoate from the organic fraction of urban waste. Sci. Total Environ. 2019, 689, 765–773. [Google Scholar] [CrossRef]
- Bosco, F.; Cirrincione, S.; Carletto, R.; Marmo, L.; Chiesa, F.; Mazzoli, R.; Pessione, E. PHA Production from Cheese Whey and “Scotta”: Comparison between a Consortium and a Pure Culture of Leuconostoc mesenteroides. Microorganisms 2021, 9, 2426. [Google Scholar] [CrossRef]
- Reddy, M.V.; Mohan, S.V. Influence of aerobic and anoxic microenvironments on polyhydroxyalkanoates (PHA) production from food waste and acidogenic effluents using aerobic consortia. Bioresour. Technol. 2012, 103, 313–321. [Google Scholar] [CrossRef]








Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Correa-Galeote, D.; Argiz, L.; Val del Rio, A.; Mosquera-Corral, A.; Juarez-Jimenez, B.; Gonzalez-Lopez, J.; Rodelas, B. Dynamics of PHA-Accumulating Bacterial Communities Fed with Lipid-Rich Liquid Effluents from Fish-Canning Industries. Polymers 2022, 14, 1396. https://0-doi-org.brum.beds.ac.uk/10.3390/polym14071396
Correa-Galeote D, Argiz L, Val del Rio A, Mosquera-Corral A, Juarez-Jimenez B, Gonzalez-Lopez J, Rodelas B. Dynamics of PHA-Accumulating Bacterial Communities Fed with Lipid-Rich Liquid Effluents from Fish-Canning Industries. Polymers. 2022; 14(7):1396. https://0-doi-org.brum.beds.ac.uk/10.3390/polym14071396
Chicago/Turabian StyleCorrea-Galeote, David, Lucia Argiz, Angeles Val del Rio, Anuska Mosquera-Corral, Belen Juarez-Jimenez, Jesus Gonzalez-Lopez, and Belen Rodelas. 2022. "Dynamics of PHA-Accumulating Bacterial Communities Fed with Lipid-Rich Liquid Effluents from Fish-Canning Industries" Polymers 14, no. 7: 1396. https://0-doi-org.brum.beds.ac.uk/10.3390/polym14071396