Estimating Above-Ground Biomass in Sub-Tropical Buffer Zone Community Forests, Nepal, Using Sentinel 2 Data
Abstract
:1. Introduction
2. Materials and Methods
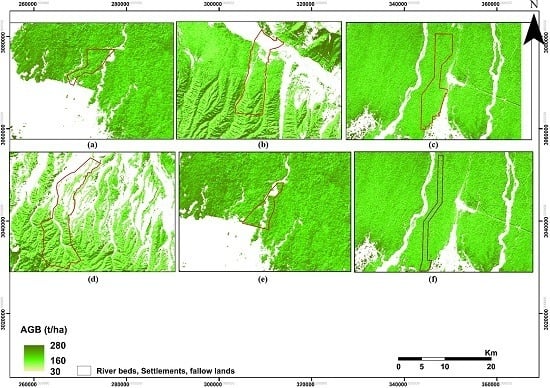
2.1. Study Area
2.2. Sampling Strategy and Field Data Collection
Estimation of Plot-Based AGB
2.3. Satellite Image Acquisition and Data Processing
2.4. Variables for Forest AGB Prediction
2.5. Statistical Analysis
3. Results
3.1. Field-Based AGB Estimates
3.2. RF Performance Based on All Variables and Important Variable
3.3. Variable Importance
3.4. AGB Mapping Using Sentinel-2 MSI Data
4. Discussion
5. Conclusions
- Sentinel-2 data effectively predicted the above-ground biomass of the sub-tropical forests, with an R2 value of 0.81 and an RMSE value of 25.57 t ha−1.
- The RF model selection of important variables did not improve the variance explained by the model (R2), but decreased the model performance error (RMSE) from 25.57 to 25.32 t ha−1.
- Technical improvements in the Sentinel-2 MSI sensor at fine medium resolution (10 and 20 m) have the potential to enable satisfactory predictions of biomass in areas of sub-tropical forest with flat terrain.
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Ebregt, A.; Greve, P.D. Buffer Zones and Their Management: Policy and Best Practices for Terrestrial Ecosystems in Developing Countries; Theme Studies Series; National Reference Centre for Nature Management: Wageningen, The Netherlands, 2000. [Google Scholar]
- Stræde, S.; Treue, T. Beyond buffer zone protection: A comparative study of park and buffer zone products’ importance to villagers living inside Royal Chitwan National Park and to villagers living in its buffer zone. J. Environ. Manag. 2006, 78, 251–267. [Google Scholar] [CrossRef] [PubMed]
- Chinembiri, T.S.; Bronsveld, M.C.; Rossiter, D.G.; Dube, T. The precision of C stock estimation in the Ludhikola watershed using model-based and design-based approaches. Nat. Res. Res. 2013, 22, 297–309. [Google Scholar] [CrossRef]
- Gara, T.; Murwira, A.; Chivhenge, E.; Dube, T.; Bangira, T. Estimating wood volume from canopy area in deciduous woodlands of Zimbabwe. South. For. 2014, 76, 237–244. [Google Scholar] [CrossRef]
- Güneralp, I.; Filippi, A.M.; Randall, J. Estimation of floodplain aboveground biomass using multispectral remote sensing and nonparametric modeling. Int. J. Appl. Earth Obs. Geoinf. 2014, 33, 119–126. [Google Scholar] [CrossRef]
- Lu, D. The potential and challenges of remote sensing-based biomass estimation. Int. J. Remote Sens. 2006, 27, 1297–1328. [Google Scholar] [CrossRef]
- Henry, M.; Picard, N.; Trotta, C.; Manlay, R.J.; Valentini, R.; Bernoux, M.; Saint-André, L. Estimating tree biomass of Sub-Saharan African forests: A review of available allometric equations. Silva Fenn. 2011, 45, 477–569. [Google Scholar] [CrossRef]
- Dong, J.; Kaufmann, R.K.; Myneni, R.B.; Tucker, C.J.; Kauppi, P.E.; Liski, J.; Buermann, W.; Alexeyev, V.; Hughes, M.K. Remote Sensing estimates of boreal and temperate forest woody biomass: Carbon pools, sources, and sinks. Remote Sens. Environ. 2003, 84, 393–410. [Google Scholar] [CrossRef]
- Chen, J.; Gu, S.; Shen, M.; Tang, Y.; Matsushita, B. Estimating aboveground biomass of grassland having a high canopy cover; an exploratory analysis of in situ hyperspectral data. Int. J. Remote Sens. 2009, 30, 6497–6515. [Google Scholar] [CrossRef]
- Dube, T.; Mutanga, O.; Elhadi, A.; Ismail, R. Intra-and-inter species biomass prediction in a plantation forest: Testing the utility of high spatial resolution space borne multispectral RapidEye sensor and advance machine learning algorithms. Remote Sens. 2014, 14, 15348–15370. [Google Scholar] [CrossRef] [PubMed]
- Dube, T.; Mutanga, O. Evaluating the utility of the medium-spatial resolution Landsat 8 multi-spectral sensor in quantifying aboveground biomass in Umgeni catchment, South Africa. ISPRS J. Photogramm. Remote Sens. 2015, 101, 36–46. [Google Scholar] [CrossRef]
- Muinonen, E.; Parikka, H.; Pokharel, Y.P.; Shrestha, S.M.; Eerikäinen, K. Utilizing a multi-source forest inventory technique, MODIS data and Landsat TM images in the production of forest cover and volume maps for the Terai Physiographic Zone in Nepal. Remote Sens. 2012, 4, 3920–3947. [Google Scholar] [CrossRef]
- Rana, P.; Korhonen, L.; Gautam, B.; Tokola, T. Effect of field plot location on estimating tropical forest above ground biomass in Nepal using airborne laser scanning data. ISPRS J. Photogramm. Remote Sens. 2014, 94, 55–62. [Google Scholar] [CrossRef]
- Shen, W.; Li, M.; Huang, C.; Wei, A. Quantifying live aboveground biomass and forest disturbance of mountainous natural and plantation forests in Northern Guangdong, China, based on Multi-temporal Landsat, PALSAR and field plot data. Remote Sens. 2016, 8, 595. [Google Scholar] [CrossRef]
- Bajcsy, P.; Groves, P. Methodology for hyperspectral band selection. Photogramm. Eng. Remote Sens. 2004, 70, 793–802. [Google Scholar] [CrossRef]
- Govender, M.; Chetty, K.; Bulcock, H. A review of hyperspectral remote sensing and its application in vegetation and water resource studies. Water S. Afr. 2007, 33, 145–152. [Google Scholar] [CrossRef]
- Lefsky, M.A.; Cohen, W.B.; Spies, T.A. An evaluation of alternative remote sensing products for forest inventory, monitoring, and mapping of Douglas-fir forests in western Oregon. Can. J. For. Res. 2001, 31, 78–87. [Google Scholar] [CrossRef]
- Plaza, A.; Benediktsson, J.A.; Boardman, J.W.; Brazile, J.; Bruzzone, L.; Camps-Valls, G.; Chanussot, J.; Fauvel, M.; Gamba, P.; Gualtieri, A.; et al. Recent advances in techniques for hyperspectral image processing. Remote Sens. Environ. 2009, 113, 110–122. [Google Scholar] [CrossRef]
- Mathieu, R.; Naidoo, L.; Cho, M.A.; Leblon, B.; Main, R.; Wessels, K.; Asner, G.P.; Buckely, J.; Aardt, J.V.; Erasmus, B.F.N.; et al. Toward structural assessment of semi-arid African savannahs and woodlands: The potential of multitemporal polarimetric RADARSAT-2 fine beam images. Remote Sens. Environ. 2013, 138, 215–231. [Google Scholar] [CrossRef]
- Sibanda, M.; Mutanga, O.; Rouget, M. Examining the potential of Sentinel-2 MSI spectral resolution in quantifying above ground biomass across different fertilizer treatments. ISPRS J. Photogramm. Remote Sens. 2015, 110, 55–65. [Google Scholar] [CrossRef]
- Gibbs, H.K.; Brown, S.; Niles, J.O.; Foley, J.A. Monitoring and estimating tropical forest carbon stocks: Making REDD a reality. Environ. Res. Lett. 2007, 2, 045023. [Google Scholar] [CrossRef]
- Hall, F.G.; Bergen, K.; Blair, J.B.; Dubayah, R.; Houghton, R.; Hurtt, G.; Kellndorfer, J.; Lefsky, M.; Ranson, J.; Saatchi, S.; et al. Characterizing 3D vegetation structure from space; mission requirements. Remote Sens. Environ. 2011, 115, 2753–2775. [Google Scholar] [CrossRef]
- Laurin, G.V.; Chen, Q.; Lindsell, J.A.; Coomes, D.A.; Del Frate, F.; Guerriero, L.; Pirotti, F.; Valentini, R. Aboveground biomass estimation in an African tropical forest with lidar and hyperspectral data. ISPRS J. Photogramm. Remote Sens. 2014, 89, 49–58. [Google Scholar] [CrossRef]
- Foody, G.M.; Boyd, D.S.; Cutler, M.E. Predictive relations of tropical forest biomass from Landsat TM data and their transferability between regions. Remote Sens. Environ. 2003, 85, 463–474. [Google Scholar] [CrossRef]
- Hall, R.J.; Skakun, R.S.; Arsenault, E.J.; Case, B.S. Modeling forest stand structure attributes using Landsat ETM+ data: Application of mapping of aboveground biomass and stand volume. For. Ecol. Manag. 2006, 225, 378–390. [Google Scholar] [CrossRef]
- Lu, D. Aboveground biomass estimation using Landsat TM data in the Brazilian Amazon. Int. J. Remote Sens. 2005, 26, 2509–2525. [Google Scholar] [CrossRef]
- Powell, S.L.; Cohen, W.B.; Healey, S.P.; Kennedy, R.E.; Moisen, G.G.; Pierce, K.B.; Ohmann, J.L. Quantification of live aboveground forest biomass dynamics with Landsat time-series and field inventory data: A comparison of empirical modeling approaches. Remote Sens. Environ. 2010, 114, 1053–1068. [Google Scholar] [CrossRef]
- Kasischke, E.S.; Goetz, S.; Hansen, M.C.; Ozdogan, M.; Rogan, J.; Ustin, S.L.; Woodcock, C.E. Remote Sensing for Natural Resource Management and Environmental Monitoring, 3rd ed.; John and Wiley and Sons, Inc.: Hoboken, NJ, USA, 2014. [Google Scholar]
- Steininger, M.K. Satellite estimation of tropical secondary forest above-ground biomass: Data from Brazil and Bolivia. Int. J. Remote Sens. 2000, 21, 1139–1157. [Google Scholar] [CrossRef]
- Broge, N.H.; Mortensen, J.V. Deriving green crop area index and canopy chlorophyll density of winter wheat from spectral reflectance data. Remote Sens. Environ. 2002, 81, 45–57. [Google Scholar] [CrossRef]
- Broge, N.H.; Leblanc, E. Comparing prediction power and stability of broadband and hyperspectral vegetation indices for estimation of green leaf area index and canopy chlorophyll density. Remote Sens. Environ. 2001, 76, 156–172. [Google Scholar] [CrossRef]
- Curran, P.J. Imaging spectrometry for ecological applications. Int. J. Appl. Earth Obs. Geoinf. 2001, 3, 305–312. [Google Scholar] [CrossRef]
- Mutanga, O.; Skidmore, A.K. Narrow band vegetation indices to overcome the saturation problem in biomass estimation. Int. J. Remote Sens. 2004, 25, 3999–4014. [Google Scholar] [CrossRef]
- Underwood, E.C.; Ustin, S.L.; Ramirez, C.M. A comparison of spatial and spectral image resolution for mapping invasive plants in coastal California. Environ. Manag. 2007, 39, 63–83. [Google Scholar] [CrossRef] [PubMed]
- Gómez, M.G.C. Joint Use of Sentinel-1 and Sentinel-2 for Land Cover Classification: A Machine Learning Approach. Master’s. Thesis, Lund University, Lund, Sweden, 2017. [Google Scholar]
- Aschbacher, J.; Milagro-Pérez, M.P. The European Earth monitoring (GMES) programme: Status and perspectives. Remote Sens. Environ. 2012, 120, 3–8. [Google Scholar] [CrossRef]
- Ramoelo, A.; Cho, M.; Mathieu, R.; Skidmore, A.K. Potential of Sentinel-2 spectral configuration to assess rangeland quality. J. Appl. Remote Sens. Environ. 2015, 124, 516–533. [Google Scholar] [CrossRef]
- Shoko, C.; Mutanga, O. Examining the strength of the newly-launched Sentinel 2 MSI sensor in detecting and discriminating subtle differences between C3 and C4 grass species. ISPRS J. Photogramm. Remote Sens. 2017, 129, 32–40. [Google Scholar] [CrossRef]
- Forkuor, G.F.; Dimobe, K.; Serme, I.; Tondoh, J.E. Landsat-8 vs. Sentinel-2: Examining the added value of Sentinel-2’s red-edge bands to land-use and land cover mapping in Burkina Faso. GISci. Remote Sens. 2017, 1–24. [Google Scholar] [CrossRef]
- Clevers, J.G.P.W.; Gitelson, A.A. Remote estimation of crop and grass chlorophyll and nitrogen content using red-edge bands on Sentinel-2 and -3. Int. J Appl. Earth Obs. Geoinf. 2012, 23, 344–351. [Google Scholar] [CrossRef]
- Kaplan, G.; Avdan, U. Mapping and monitoring wetlands using Sentinel-2 satellite imagery. ISPRS Ann. Photogramm. Remote Sens. Spat. Inf. Sci. 2017, IV-4/W4, 271–277. [Google Scholar] [CrossRef]
- Godinho, S.; Guiomar, N.; Gil, A. Estimating tree canopy percentage in Mediterranean slivopastoral systems in suing Sentinel-2A imagery and the stochastic gradient boosting algorithm. Int. J. Remote Sens. 2017, 1–23. [Google Scholar] [CrossRef]
- Karna, Y.K.; Hussin, Y.A.; Gilani, H.; Bronsveld, M.C.; Murthy, M.S.R.; Qamer, F.M.; Karky, B.S.; Bhattarai, T.; Aigong, X.; Baniya, C.B. Integration of WorldView-2 and airborne LiDAR data for tree species level carbon stock mapping in Kayar Khola watershed, Nepal. Int. J. Appl. Earth Obs. Geoinf. 2015, 38, 280–291. [Google Scholar] [CrossRef]
- Baral, S. Mapping Carbon Stock Using High Resolution Satellite Images in Sub-Tropical Forest of Nepal. Ph.D. Thesis, University of Twente, Enschede, The Netherlands, 2011. [Google Scholar]
- FRA/DFRS. Terai Forests of Nepal (2010-2012); Forest Resource Assessment Nepal/Department of Forest Research and Survey: Babarmahal, Kathmandu, Nepal, 2014.
- Murthy, M.S.R.; Wesselman, S.; Gilani, H. Multi-Scale Forest Biomass Assessment and Monitoring in the Hindu Kush Himalayan Region: A Geospatial Perspective; International Centre for Integrated Mountain Development (ICIMOD): Kathmandu, Nepal, 2015; pp. 70–82. [Google Scholar]
- Strobl, C.; Boulesteix, A.L.; Kneib, T.; Augustin, T.; Zeileis, A. Conditional variable importance for random forests. BMC Bioinf. 2008, 9, 307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karlson, M.; Ostwald, M.; Reese, H.; Sanou, J.; Tankoano, B.; Mattsson, E. Mapping tree canopy cover and aboveground biomass in Sudano-Sahelian Woodlands using Landsat 8 and Random forest. Remote Sens. 2015, 7, 10017–10041. [Google Scholar] [CrossRef]
- Dunne, K.; Cunningham, P.; Azuaje, F. Solution to instability problems with sequential wrapper-based approaches to feature selection. J. Mach. Learn. Res. 2002, 1–22. Available online: https://www.scss.tcd.ie/publications/tech-reports/reports.02/TCD-CS-2002-28.pdf (accessed on 27 December 2017).
- Saeys, Y.; Inza, I.; Larrañaga, P. A review of feature selection techniques in bioinformatics. Bioinformatics 2007, 23, 2507–2517. [Google Scholar] [CrossRef] [PubMed]
- Chave, J.; Andalo, C.; Brown, S.; Cairns, M.A.; Chambers, J.Q.; Eamus, D.; Fölster, H.; Fromard, F.; Higuchi, N.; Kira, T.; et al. Tree allometry and improved estimation of carbon stocks and balance of tropical forest. Ecosyst. Ecol. 2005, 145, 87–99. [Google Scholar] [CrossRef] [PubMed]
- Chaturvedi, A.N.; Khanna, L.S. Forest Mensuration; International Book Distributors: Dehra Dun, India, 1982. [Google Scholar]
- Chavez, P.S. An improved-dark object subtraction technique for atmospheric scattering correction of multispectral data. Remote Sens. Environ. 1988, 24, 459–479. [Google Scholar] [CrossRef]
- Frampton, W.J.; Dash, J.; Watmough, G.; Milton, J.M. Evaluating the capabilities of Sentinel-2 for quantitative estimation of biophysical variables in vegetation. ISPRS J. Photogramm. Remote Sens. 2013, 82, 83–92. [Google Scholar] [CrossRef]
- Stagakis, S.; Markos, N.; Sykioti, O.; Kyparissis, A. Monitoring canopy biophysical and biochemical parameters in ecosystem scale using satellite hyperspectral imagery: An application on a Phlomis fruticosa Mediterranean ecosystem using multiangular CHRIS/PROBA observations. Remote Sens. Environ. 2010, 114, 977–994. [Google Scholar] [CrossRef]
- Tucker, C.J. Red and Photographic infrared linear combination for monitoring vegetation. Remote Sens. Environ. 1979, 8, 127–150. [Google Scholar] [CrossRef]
- Sims, D.A.; Gamon, J.A. Relationships between leaf pigment content and spectral reflectance across a wide range of species, leaf structures and development stages. Remote Sens. Environ. 2002, 81, 337–354. [Google Scholar] [CrossRef]
- Huete, A.; Didan, K.; Miura, T.; Rodriguez, E.P.; Gao, X.; Ferreira, L.G. Overview of the radiometric and biophysical performance of the MODIS vegetation indices. Remote Sens. Environ. 2002, 83, 195–213. [Google Scholar] [CrossRef]
- Jordan, C.F. Derivation of leaf-area index from quality of light on the forest floor. Ecology 1969, 50, 663–666. [Google Scholar] [CrossRef]
- Hill, M.J. Vegetation index suites as indicators of vegetation state in grassland and savanna: An analysis with simulated SENTINEL 2 data for a North American transect. Remote Sens. Environ. 2013, 137, 94–111. [Google Scholar] [CrossRef]
- Merzlyak, M.N.; Gitelson, A.A.; Chivkunova, O.B.; Rakitin, V.Y. Non-destructive optical detection of pigment changes during leaf senescence and fruits ripening. Physiol. Plant. 1999, 106, 135–141. [Google Scholar] [CrossRef]
- Hardisky, M.A.; Klemas, V.; Smart, R.M. The influence of soil salinity, growth form, and leaf moisture on the spectral radiance of Spartina alterniflora canopies. Photogramm. Eng. Remote Sens. 1983, 49, 77–83. [Google Scholar]
- Chen, J.C.; Yang, C.M.; Wu, S.T.; Chung, Y.L.; Charles, A.L.; Chen, C.T. Leaf chlorophyll content and surface spectral reflectance of tree species along a terrain gradient in Taiwan’s Kenting National Park. Bot. Stud. 2007, 48, 71–77. [Google Scholar]
- Huete, A.R. A soil-adjusted vegetation index (SAVI). Remote Sens. Environ. 1988, 25, 295–309. [Google Scholar] [CrossRef]
- Kross, A.; McNairn, H.; Lapen, D.; Sunohara, M.; Champagne, C. Assessment of RapidEye vegetation indices for estimation of leaf area index and biomass in corn and soybean crops. Int. J. Appl. Earth Obs. Geoinf. 2015, 34, 235–248. [Google Scholar] [CrossRef]
- Gitelson, A.; Merzlyak, M.N. Spectral reflectance changes associated with autumn senescence of Aesculus hippocastanum L. and Acer plantanoides L. leaves. Spectral features and relation to chlorophyll estimation. J. Plant Physiol. 1994, 143, 286–292. [Google Scholar] [CrossRef]
- Sharma, L.K.; Bu, H.; Denton, A.; Franzen, D.W. Active-optical sensors using red NDVI compared to red edge NDVI for prediction of corn grain yield in North Dakota, USA. Sensors 2015, 15, 27832–27853. [Google Scholar] [CrossRef] [PubMed]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Prasad, A.M.; Iverson, L.R.; Liaw, A. Newer classification and regression tree techniques: Bagging and random forests for ecological prediction. Ecosystems 2006, 9, 181–199. [Google Scholar] [CrossRef]
- Ozcift, A. Random forest ensemble classifier trained with data resampling strategy to improve cardiac arrhythmia diagnosis. Comput. Biol. Med. 2011, 41, 265–271. [Google Scholar] [CrossRef] [PubMed]
- Palmer, D.S.; O’Boyle, N.M.; Glen, R.C.; Mitchell, J.B.O. Random forest model to predict aqueous solubility. J. Chem. Inf. Model. 2007, 47, 150–158. [Google Scholar] [CrossRef] [PubMed]
- Reif, D.M.; Motsinger, A.A.; McKinney, B.A.; Crowe, J.E.; Moore, J.H. Feature selection using a random forests classifier for the integrated analysis of multiple data types. IEEE Symp. Comput. Intell. Bioinf. Comput. Biol. 2006, 1–8. [Google Scholar] [CrossRef]
- Adam, E.; Mutanga, O.; Rugege, D. Multispectral and hyperspectral remote sensing for identification and mapping of wetland vegetation: A review. Wetlands Ecol. Manag. 2010, 18, 281–296. [Google Scholar] [CrossRef]
- Muukkonen, P.; Heiskanen, J. Estimating biomass for boreal forest using ASTER satellite data combined with standwise forest inventory data. Remote Sens. Environ. 2005, 99, 434–447. [Google Scholar] [CrossRef]
- Adan, M.S. Integrating Sentinel-2 Derived Vegetation Indices and Terrestrial Laser Scanner to Estimate Above-Ground Biomass/Carbon in Ayer Hitam Tropical Forest Malaysia. Master’s Thesis, The University of Twente, Enschede, The Netherlands, 2017. [Google Scholar]
- Laurin, G.V.; Puletti, N.; Hawthorne, W.; Liesenberg, V.; Corona, P.; Papale, D.; Chen, Q.; Valentini, R. Discrimination of Tropical forest types, dominant species, and mapping of functional guilds by hyperspectral and stimulated multispectral Sentinel-2 data. Remote Sens. Environ. 2016, 176, 163–176. [Google Scholar] [CrossRef] [Green Version]
- Gómez, C.; White, J.C.; Wulder, M.A. Optical remotely sensed time series data for land cover classification: A review. ISPRS J. Photogramm. Remote Sens. 2016, 116, 55–72. [Google Scholar] [CrossRef]
- Adam, E.; Mutanga, O.; Abdel-Rahman, E.M.; Ismail, R. Estimating standing biomass in papyrus (Cyperus papyrus L.) swamp: Exploratory of in situ hyperspectral indices and random forest regression. Int. J. Remote Sens. 2014, 35, 693–714. [Google Scholar] [CrossRef]
- Liu, K.; Wang, J.; Zeng, W.; Song, J. Comparison and evaluation of three models for estimating forest above ground biomass using TM and GLAS data. Remote Sens. 2017, 9, 341. [Google Scholar] [CrossRef]
- Dube, T.; Mutanga, O. Investigating the robustness of the newly Landsat-8 Operational Land Imager derived texture metrics in estimating plantation forest aboveground biomass. ISPRS J. Photogramm. Remote Sens. 2015, 108, 12–32. [Google Scholar] [CrossRef]
- Wu, C.; Shen, H.; Wang, K.; Shen, A.; Deng, J.; Gan, M. Landsat Imagery-based aboveground biomass estimation and change investigation related to human activities. Sustainability 2016, 8, 159. [Google Scholar] [CrossRef]
- Mutanga, O.; Adam, E.; Cho, M.A. High density biomass estimation for wetland vegetation using Worldview-2 imagery and random forest regression algorithm. Int. J. Appl. Earth Obs. Geoinf. 2012, 18, 399–406. [Google Scholar] [CrossRef]





| Designation | Sentinel-2 Bands | |
|---|---|---|
| Spectral: | ||
| Blue | B2 | |
| Green | B3 | |
| Red | B4 | |
| RE 1 | B5 | |
| RE 2 | B6 | |
| RE 3 | B7 | |
| NIR | B8 | |
| RE 4 | B8a | |
| SWIR1 | B11 | |
| SWIR2 | B12 | |
| Vegetation Indices | Equations | References |
| NDVI | (NIR − R/NIR + R) | [56] |
| RGR | Red665/Green560 | [57] |
| EVI | 2.5*((NIR − R)/ (1 + NIR + 6R − 7.5 Blue)) | [58] |
| SR | NIR/RED | [59] |
| PSRI | (ꝭ665 − ꝭ560/ꝭ740) | [60,61] |
| NDII | (ꝭ842 − ꝭ1610/ꝭ842 + ꝭ1610) | [62] |
| RE NDVI | ꝭ842 − ꝭ740/ꝭ842 + ꝭ740 | [63] |
| SAVI | (NIR − R)/(NIR + R + L)*1.5 | [64] |
| IRECI | NIR − R/(RE1/RE2) | [54] |
| S2REP | [705 + 35*(0.5*(B7 + B4)/2) − B5)/(B6 − B5)] | [54] |
| Red-edge-based NDVIs: | ||
| 1 | (NIR − RE1/NIR + RE1) | [65] |
| 2 | (NIR − RE2/NIR + RE2) | [65,66] |
| 3 | (NIR − RE3/NIR + RE3) | [67] |
| 4 | (NIR − RE4/NIR + RE4) | [65] |
| No. | Name of Forest | Total No. of Plots | Min (t ha−1) | Max (t ha−1) | Mean (t ha−1) | Standard Deviation |
|---|---|---|---|---|---|---|
| 1 | Ratomate Deurali BZCF | 17 | 35.25 | 115.95 | 115.87 | 33.66 |
| 2 | Jyamire BZCF | 30 | 34.75 | 304.19 | 166.81 | 88.60 |
| 3 | Radha Krishna BZCF | 59 | 105.40 | 230.72 | 174.91 | 35.51 |
| 4 | Janahit BZCF | 30 | 96.95 | 286.56 | 172.18 | 47.32 |
| 5 | Shrijana BZCF | 21 | 30.34 | 200.22 | 117.94 | 56.33 |
| 6 | Mushaharnimae BZCF | 16 | 90.95 | 210.48 | 170.52 | 40.26 |
| No. of Variables Used | Eliminated Variable (Backward) | R2 | RMSE | relRMSE |
|---|---|---|---|---|
| 24 | Full predictors | 0.81 | 25.57 | 15.99 |
| 23 | SAVI | 0.81 | 25.45 | 15.91 |
| 22 | Band 4 | 0.81 | 25.69 | 16.06 |
| 21 | PSRI | 0.81 | 25.36 | 15.86 |
| 20 | Band8 | 0.81 | 25.32 | 15.84 |
| 19 | IRECI | 0.80 | 25.79 | 16.13 |
| 18 | Band11 | 0.81 | 25.48 | 15.94 |
| 17 | EVI | 0.80 | 25.72 | 16.08 |
| 16 | RE_NDVI2 | 0.8 | 25.89 | 16.19 |
| 15 | Band12 | 0.80 | 26.01 | 16.27 |
| 14 | RGR | 0.80 | 25.84 | 16.16 |
| 13 | RE_NDVI | 0.80 | 25.97 | 16.24 |
| 12 | Band5 | 0.79 | 26.21 | 16.39 |
| 11 | Band3 | 0.79 | 26.45 | 16.54 |
| 10 | RE_NDVI4 | 0.79 | 26.19 | 16.38 |
| 9 | NDII | 0.79 | 26.56 | 16.61 |
| 8 | RE_NDVI3 | 0.76 | 28.08 | 17.56 |
| 7 | S2REP | 0.75 | 28.44 | 17.79 |
| 6 | Band2 | 0.73 | 29.55 | 18.48 |
| 5 | RE_NDVI1 | 0.71 | 30.6 | 19.2 |
| 4 | SR | 0.71 | 30.58 | 19.13 |
| 3 | NDVI | 0.70 | 31.80 | 19.88 |
| 2 | Band6 | 0.70 | 31.80 | 19.88 |
| 1 | Band7 | 0.57 | 37.58 | 23.50 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pandit, S.; Tsuyuki, S.; Dube, T. Estimating Above-Ground Biomass in Sub-Tropical Buffer Zone Community Forests, Nepal, Using Sentinel 2 Data. Remote Sens. 2018, 10, 601. https://0-doi-org.brum.beds.ac.uk/10.3390/rs10040601
Pandit S, Tsuyuki S, Dube T. Estimating Above-Ground Biomass in Sub-Tropical Buffer Zone Community Forests, Nepal, Using Sentinel 2 Data. Remote Sensing. 2018; 10(4):601. https://0-doi-org.brum.beds.ac.uk/10.3390/rs10040601
Chicago/Turabian StylePandit, Santa, Satoshi Tsuyuki, and Timothy Dube. 2018. "Estimating Above-Ground Biomass in Sub-Tropical Buffer Zone Community Forests, Nepal, Using Sentinel 2 Data" Remote Sensing 10, no. 4: 601. https://0-doi-org.brum.beds.ac.uk/10.3390/rs10040601