1. Introduction
As one of the main terrestrial ecosystems, forests play a vital role in the conservation of biological diversity and suppression of climate change. Forest inventory is the systematic collection of data and forest information needed for assessment or analysis for the sake of forest management and ecosystem sustaining. The comprehensive information about the state and dynamics of forests, such as tree species and their distribution, number of trees per acre, basal area, timber volume, increment of timber volume and mean tree height, can be acquired through the statistical forest inventory [
1]. Traditionally, forest inventory is labor-intensive, time-consuming and restricted by spatial accessibility because most of the forest attributes and variables are estimated by measuring sample plots manually in field surveys [
2].
In recent years, many efforts have been made to decrease the costs by developing inventory systems that are based on remote sensing technologies. Intensive studies have been focused on optical aerial photography interpretation. However, large-scale aerial photos or high-spatial resolution remotely-sensed imagery do not directly provide three-dimensional (3D) forest structural information [
3]. This limitation can be overcome by a technology called airborne LiDAR (Light Detection And Ranging) [
4]. Airborne LiDAR employs state of the art laser technology coupled with high-end GPS (Global Positioning System) and IMU (Inertial Measurement Unit) systems as a means of geo-referencing to produce accurate and detailed scan data from an airborne platform [
5]. Compared with passive imaging, LiDAR has the advantage of directly measuring the 3D coordinates of forest canopies [
3]. Furthermore, considering that the laser beam emitted from the airborne LiDAR is able to penetrate beneath forest canopies down to the ground, the LiDAR can provide an unstructured 3D point cloud that is a discrete model of the forest [
6]. Due to its ability to generate 3D data with high spatial resolution and accuracy, the airborne LiDAR technology is being increasingly used in remote sensing-based forest resource monitoring [
4].
Airborne LiDAR-based remote sensing for forest inventory applications necessitates proper extraction of individual trees and accurate delineation of their canopy structure. Once a single tree has been detected from the remotely-sensed data (namely the point cloud generated by the LiDAR), structural attributes such as tree height, crown diameter, canopy-based height, basal area, Diameter at Breast Height (DBH), wood volume, biomass and species type can be derived [
4]. A variety of methods has been proposed to identify individual trees or tree crowns using airborne LiDAR data, which fall into one of the two categories: raster-based approaches or point-based approaches. Raster-based delineation utilizes the Canopy Height Model (CHM) derived from the raw LiDAR data, which is the difference between the canopy surface height and a Digital Elevation Model (DEM) of the Earth’s surface [
3]. In a typical scenario that adopts a CHM-based method, a local maximum filter is used to detect treetops, and then, the individual tree crowns are delineated with a marker-controlled watershed segmentation scheme, a region growing algorithm or a pouring algorithm [
3,
7,
8,
9,
10,
11]. The spatial 2D wavelet analysis is also applied to automatically determine the location, height and crown diameter of individual trees from the CHM in some raster-based scenarios [
12]. As the CHM is essentially a raster image interpolated from spatially-discrete points depicting the vegetation canopy top, there can be inherent errors and uncertainties in it [
4]. The spatial errors introduced in the interpolation process could decrease the accuracy of tree segmentations and relevant measurements [
13]. The process of producing a CHM, preserving just return data from the surface, could also lead to loss of information from different levels in the canopy, and thus, a further reduction in the data available for individual tree delineation [
14]. Generally, CHM-based methods tend to merge crowns in dense stands of deciduous trees [
10] or fail to detect understory trees in a multi-layered forest [
15]. Instead, point-based approaches work directly on the original point cloud data rather than rasterized images. Since direct processing on the LiDAR’s raw data can avoid errors introduced by interpolation, it is generally acknowledged that the point-based methods outperform CHM-based methods [
14].
Region growing, as a category of well-known image segmentation methods [
16], has been frequently used to delineate individual trees based on the original LiDAR point data [
2,
4,
17]. In general, region growing-based methods are easily implemented, but they are not particularly robust as the segmentation quality strongly depends on both multiple criteria and the selection of seed points/regions, where no universally-valid criterion exists [
18]. Segmenting individual trees directly from the original LiDAR data can also resort to clustering-based approaches. One of the most popular clustering methods is the
k-means algorithm. The basic
k-means clustering scheme or more complicated mechanisms based on a certain modified
k-means algorithm were employed to extract individual trees from the point clouds with seed points extracted from a rasterized Digital Surface Model (DSM) by a local maximum filter [
19,
20]. The
k-means clustering is so simple as to be easy to implement and apply, even on large datasets; however, one of its biggest drawbacks is that
k, the number of clusters, is a predefined parameter, and an inappropriate choice of
k may yield poor clustering results [
21]. Besides, the performance of the
k-means clustering is heavily dependent on the locations of starting locations or seed points, which are most often randomly chosen from the entire dataset and used as the initial centroids [
22]. A crucial issue in applying the above-mentioned region growing or
k-means clustering approaches is how to determine the seed points, whose numbers and locations will strongly influence subsequent processing steps. In the scenario of isolating and delineating individual trees, the seed points are usually considered to be the estimated treetops, and the region growing algorithms utilize these seed points to grow regions or tree crowns around them until some stop criteria are satisfied, while the
k-means clustering starts an iterative procedure with the seed points as initial means of all points [
17]. However, it is not a trivial problem to detect treetops for canopies accurately. The common method to identify treetops is to extract local maxima using local maximum filtering with variable window sizes in a rasterized digital model such as CHM or the Canopy Maxima Model (CMM), or to find the highest point for a particular tree crown in the point dataset with an adaptive search radius when raw LiDAR data are used [
3,
9]. Yet, it is very difficult to determine an appropriate size for the moving window or search radius to detect the treetops because different sizes should be applied to different regions for optimal detection accuracy [
2].
As a powerful and versatile feature-space analysis technique, mean shift has found much interest in clustering of LiDAR data for forest inventory applications. Mean shift-based schemes for clustering data points with similar vegetation modes together, which directly work on the 3D point cloud acquired with a small-footprint LiDAR, were reported in [
6,
23]. Ferraz et al. proposed a multi-scale mean shift technique where modes were computed with a vegetation stratum-dependent kernel bandwidth to simultaneously segment vertical and horizontal structures of forest canopies and decompose the entire point cloud into 3D clusters that correspond to individual tree crowns [
15,
24]. Compared with other kinds of clustering approaches (e.g., region growing or
k-means clustering), the mean shift is robust to initializations and can handle arbitrarily-shaped clusters, as it does not depend on any geometric model assumptions. More importantly, the mean shift-based approaches outperform other kinds of methods in detecting small or suppressed trees in the understory [
24]. However, because it is sensitive to the selection of kernel bandwidth, the mean shift requires the bandwidth parameter to be tuned reasonably well for performance optimization, and thus, the bandwidth calibration is actually a major challenge for the mean shift-based schemes. A self-adaptive method calibrating the kernel bandwidth as a function of a local tree allometric model that can adapt to the local forest structure was developed by Ferraz et al. in [
25]. Our previous study proposed an adaptive mean shift-based clustering approach to segment the 3D forest point clouds and identify individual tree crowns, where much effort was put into the adaptive bandwidth settings: the kernel sizes are automatically changed according to the estimated crown diameters of distinct trees [
26]. This approach has been tested over a multi-layered coniferous and broad-leaved mixed forest located in South China: the overall tree detection rate reaches 86% (92% of the detected trees are correct), and a specific detection rate of 48% was observed for the suppressed trees (the “precision” is 67%) [
26]. It should be emphasized that nearly 30% more suppressed trees can be identified by this adaptive approach than the one using a fixed kernel bandwidth [
26].
Although the adaptive mean shift clustering can produce relatively high segmentation accuracies when compared with a fixed bandwidth one, there are still obvious omission and commission errors (or under- and over-segmentations) in the ultimate detection results. This is especially the case for a structurally complex forest, which often has a multi-layered structure and consists of trees occluding each other. Due to overlapping point cloud patches caused by complex terrain, closed forest canopies or other reasons, groups of trees standing very close to each other, identified as several single trees in the field inventory, might be treated as one cluster composed of all of these trees while performing the mean shift segmentation over the LiDAR point cloud. Besides, in a forest with multiple canopy layers, many trees have complex crown structures, the tree crowns have a great range of sizes and their branches interlace with each other or extend out so that they look like trees. This will cause false detection and split a large tree into several small trees, which leads to omission and commission errors of tree identification. The errors are mainly caused by the top-down segmentation scheme in which individual tree segmentation mostly relies on accurate characterization of tree crowns; however, in fact, the tree crowns are difficult to identify and delineate from the forest canopy with a complex structure. Higher accuracies of tree detections could be achieved by adopting a bottom-up segmentation strategy. Reitberger et al. used Random Sample Consensus (RANSAC)-based 3D line adjustment to detect tree trunks (or stems) and a normalized cut method to isolate trees [
11]. A bottom-up method was proposed in [
27] to extract and segment tree trunks based on the intensity and topological relationships of the points and then allocate other points to exact tree trunks according to 2D and 3D distances. Shendryk et al. developed a two-step, bottom-up methodology for individual tree delineation: firstly, identify individual tree trunks based on conditional Euclidean distance clustering, and then, delineate tree crowns using an algorithm based on random walks in the graph paradigm [
28]. Essentially, the key idea of a bottom-up strategy is to identify the trunk points of individual trees and use the thus detected trunks as seeds for further crown delineation.
This study aims to solve the challenges of the mean shift-based tree segmentation in structurally complex forests by combining conventional clustering with a bottom-up strategy. We focus on developing an improved technological scheme using a trunk detection-aided adaptive mean shift algorithm for accuracy refinement to segment individual trees and estimate tree structural parameters based solely on the airborne LiDAR data. The main difference between this paper and our previous study, or the major contribution of this study, is the introduction of the tree trunk detection technique into the adaptive mean shift-based clustering scheme. The detected trunk information can not only be utilized to adaptively calibrate the kernel bandwidth of the mean shift procedure, but also be combined with mean shift clustering results to perform final detection of individual trees. The objectives of this study are (i) to highlight a new adaptive mean shift-based clustering approach aided by tree trunk detection using airborne LiDAR data for individual tree detection and tree-based parameter estimation, (ii) to present the results of the proposed approach when applied to small-footprint LiDAR data acquired in a field survey performed in a multi-layered evergreen mixed forest located in South China and (iii) to evaluate the new approach with respect to the tree detection rates and parameter estimation accuracy.
3. Method
3.1. Adaptive Mean Shift-Based Clustering
In forestry applications, a LiDAR point cloud can be treated as an empirical multimodal distribution where each mode, defined as a local maximum both in height and density, corresponds to a tree top, and then, each point in the 3D dataset can be considered as sampled from the underlying probability density function. Feature vectors extracted from the original point cloud data constitute a huge feature space, on which the mean shift-based clustering analysis is performed. Mean shift is a powerful non-parametric feature-space analysis technique that can be used for many purposes like mode finding, pattern recognition, tracking, filtering, clustering, etc. Its original idea was firstly introduced by Fukunaga and Hostetler [
32] and has been extended to be applicable in fields like image and video processing, computer vision and machine learning [
33,
34]. Mean shift’s uses in LiDAR data processing were firstly explored by Melzer in [
35] and have gradually become an effective tool for filtering, classification and segmentation of unstructured point clouds.
Mean shift is an iterative algorithm using a non-parametric probability density estimator based on the Parzen window kernel function [
34]. When used for clustering of unstructured data, the mean shift algorithm aims to move each data point to the densest area in its vicinity until convergence to the local maxima by iteratively performing the shift operation based on kernel density estimation.
Figure 3 shows the density estimation procedure with the mean shift vector, which is the difference between the weighted mean and the center of a kernel window (i.e., the region of interest). Given
n data points
in the
d-dimensional space
, the mean shift vector at point
is defined as:
where
h is the bandwidth parameter and
is called the profile of the kernel
. By setting:
where
t denotes the iteration number, the iterative process converge towards the local maxima. Obviously, a mean shift vector always points toward the direction of the maximum increase in density, and successive vectors can define a path leading to a mode of the estimated density. All data points that have converged to the same mode are grouped together as a cluster. In mean shift theory, a cluster is defined as an area of higher density than the remainder of the dataset, and a dense region in feature space corresponds to a mode (or a local maximum) of the probability density distribution. The ultimate result of the mean shift procedure will associate each point in 3D space with a particular cluster.
For the mean shift algorithm, determination of the kernel bandwidth, the only parameter that needs to be specified, is very critical. It is justified that an adaptive mean shift algorithm automatically calibrating the kernel bandwidth according to local structure information outperforms that using a single bandwidth for the whole feature space. The adaptive mean shift-based clustering scheme presented in our previous study aims to decompose the 3D forest point clouds into segments corresponding to individual tree crowns [
26]. It is a two-stage sequential segmentation procedure chain: coarse segmentation (single-scale mean shift-based area partitioning) and fine segmentation (adaptive mean shift-based clustering). The highlight of the adaptive mean shift-based approach is a computationally-efficient method to dynamically calibrate the kernel bandwidth, which is not set to a fixed value or determined as a multi-scale function, but can be varied in a manner that reflects the quantitative information about the local canopy structure. Please refer to [
26] for more detailed information about this approach.
The major problem encountered when using mean shift clustering techniques to detect trees is omission and commission errors. The omission errors result from under-segmentation, which implies assigning a LiDAR point cluster to several actual trees, whereas over-segmentation (a single tree segmented into more than one LiDAR clusters) of tree crowns will lead to commission errors. Under-segmentations are more prone to happen than over-segmentations since the tree crown delineation methods based on clustering techniques have the potential of missing field-measured trees: smaller trees are the intermediate and lower height level are unable to be recognized or groups of trees with crowns overlapping each other due to short mutual distances might be treated as one LiDAR segment, in which it is difficult to distinguish and separate individual trees by a clustering-based segmentation technique.
3.2. Concept and Workflow
The adaptive mean shift-based clustering technique essentially segments the forest point clouds and delineates individual tree crowns according to the LiDAR point density distribution in 3D space due to the fact that the density of received laser pulses is high at the center of a tree crown and decreases towards the edge of the crown. It performs well in forests or forest layers with regular forms, but its detection accuracy degrades seriously when dealing with forest environments with complex patterns. The main cause is the inadequate tree finding capability inherent in the mean shift-based technique. As a top-down segmentation scheme, it works over the point cloud derived from the laser pulses, mainly sampling the forest canopy structure, but rarely takes into account the LiDAR points under individual tree canopies, which contain structural information reflecting the spatial characteristics of tree trunks. It is advantageous to add a bottom-up strategy into the methodology: the trunk-related information should be utilized to aid in segmentation and delineation of individual trees to improve overall performance.
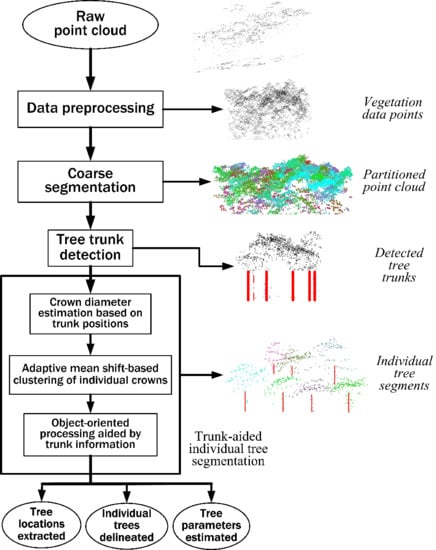
Taking into account all necessary mathematical processing used in this study, we present a flowchart of methods used in this study, as illustrated in
Figure 4, where the ellipses represent data or parameters and the rectangles represent processes or calculations. Each processing step will be described in detail in the following part of this section.
3.3. Data Preprocessing
To focus on individual tree’s delineation and measurement, we need to delete the LiDAR points belonging to the ground surface and the undersized vegetation. Considering that the ground surface is not flat and our proposed approach is based on the relative spacing of each point, we also need to remove the slope effect. First, the raw LiDAR point data were classified into ground points and above-ground points using TerraScan [
36]. The aboveground or vegetation points were used in this study. A Digital Terrain Model (DTM) was generated by interpolating the ground points using ordinary kriging, and the vegetation point cloud was normalized by subtracting the DTM height from the raw LiDAR point cloud. After the normalization process, the elevation value (namely the coordinate in the
z-dimension) of a point indicates the vertical distance from the ground to the point. Next, data points belonging to non-canopy features in the undersized vegetation strata were cleared. Points whose heights fall in a predefined range
(
is a preset threshold height) were recognized and then removed from the vegetation point cloud. In this study, we deleted all points belonging to the undersized vegetation up to a height value of 1 m from the ground level.
3.4. Coarse Segmentation
The coarse segmentation aims to partition the 3D space over each forest plot into many small irregular spatial regions, called “partitions” here, each of which contains 3D spatial features representing a single tree or a group of several actual trees. As the sizes of these segments are much smaller compared to that of the original test plot, the subsets of data points in the segments are much easier to handle than the whole dataset contained in a point cloud. For example, when much more complicated analyses and calculations are performed on these segments, which implies more complex mathematics, the total computational load could be reduced to an affordable range. We adopt the single-scale mean shift-based algorithm described in [
26] to accomplish coarse segmentation of the LiDAR point cloud. Firstly, a fast mean shift procedure using the simplest kernel function, the uniform kernel, is performed on a part of the dataset (namely the intermediate pulse data) to yield a number of point clusters, each of which reflects the spatial structure of one or more trees. Next, the first pulse data depicting the canopy surfaces are added to expand these clusters to the whole dataset by means of a so-called small square grouping technique. An example of the coarse segmentation results for a small area (10 m × 10 m) selected from a representative subplot is given in
Figure 5. For a detailed description of this processing stage, please refer to [
26].
3.5. Tree Trunk Detection
The irregular segmented region (a partition of the 3D point cloud data in a forest area of interest) resulting from the coarse segmentation process mentioned above, where obvious under-segmentations occur, usually contains several actual trees. Thus, each individual tree should be further extracted from the partition in the subsequent processing stage. However, only using a crown-based tree delineation method (e.g., the CHM-based watershed segmentation or the mean shift clustering) would result in omission errors (under-detection of trees), or commission errors (over-detection of trees), or both, for the reason that a crown segmentation mostly relies on a detection of tree tops based on local maxima or high densities, but not on a tree trunk detection. A group of trees with LiDAR point patches overlapped cannot be clearly separated by the crown-based segmentation method, whereas their trunks might be isolated from each other using points lying below tree crowns. Apparently, the introduction of the tree trunk detection would help to improve the overall tree detection rate.
The tree trunk detection adopted in this study works in a two-step algorithm. The first step aims to detach all potential crown points within a specific partition and separate out the trunk section. This goal is achieved by analyzing a vertical histogram of the points within a partition, whose idea is similar to the method presented in [
11,
37], but easier to implement (
Figure 6). Firstly, divide LiDAR points between
and
(
and
denote the minimum and maximum elevation values for all points in a partition, respectively) into
horizontal layers. Next, generate the histogram of the point distribution along the vertical direction by calculating the number of points per layer and normalizing the results by dividing each of them by the total number of points in the partition. Finally, locate the separating plane that detaches all potential crown points from the trunk section by searching the lowest horizontal layer where the point density exceeds a predefined threshold value:
.
In the second step, the separated trunk points are clustered according to their spatial neighborhood relationships to get an estimation of the number and positions of tree trunks within a specific partition and assign point groups to the estimated trunks. It is assumed that several points that are spatially close together will form a trunk. Isolated points with no or just a few neighbors are assumed to be noise or sparse scrubby vegetation. Due to not knowing the number of trunks a priori, the mean shift algorithm (a non-parametric clustering technique) is also adopted in this step. Prior to cluster analysis, the trunk points are all projected onto a horizontal plane, and a simple mean shift clustering procedure is then performed in the 2D space constituted by these projected points using a flat kernel function:
where
denotes the data point in the 2D Euclidean space and
is the kernel bandwidth or search window size. Considering that the DBH of the trees in the study area rarely exceeds 0.5 m, we assign a fixed value of 0.75 m (1.5 × 0.5 m) to the bandwidth
. Any cluster with more than 5 data points is deemed to be a tree trunk; otherwise, it should be treated as noisy data or a segment of lower shrubs or weeds and would be deleted from the trunk points. The approximate trunk position is estimated by finding a cluster’s stationary point.
Figure 7 shows the results of tree trunk detection using the above-described mean shift clustering for a typical partition. As can be seen from this example, an obvious commission error, appearing as a falsely-detected trunk, resulted from this tree trunk detection technique.
3.6. Trunk-Aided Mean Shift Clustering for Segmentation of Individual Trees
The aim of this stage is to segment each individual tree from the LiDAR point cloud for a particular forest partition by performing an adaptive mean shift-based clustering analysis with the aid of tree trunk detection results. Although the results contain some errors, they would indicate the general horizontal distribution of trees in a partition, reflecting the basic horizontal structure in a forest area of interest. From this useful information about the local vegetation structure, we can get an estimate of the individual tree crown size, which in turn can be utilized to calibrate the kernel bandwidth of the mean shift procedure adaptively. More importantly, the trunk detection results can provide supplementary information with which the point clusters generated by the adaptive mean shift algorithm are combined to perform the final detection of individual trees. The combinational use of mean shift clustering results and detected tree trunk information in this stage would serve as references against each other to reduce over- and under-segmentations and thus help to improve the overall tree detection accuracy.
The adaptive mean shift procedure requires the kernel bandwidth to be automatically calibrated according to specific tree crown sizes. However, acquisition of the structural data reflecting the local forest canopy relies on the accurate extraction of individual trees from the LiDAR point clouds. Therefore, this is a chicken and egg debate, and it is very difficult to obtain exact quantitative information about the local spatial structures of the forest canopy prior to complete clustering of individual trees. Yet, it is possible to get rough information about the sizes of individual tree crowns ahead of a tree delineation process, which provides a basis for adaptive determination of the kernel bandwidth. In this study, the data of tree crown widths are roughly estimated based on tree trunk detection results. The estimation algorithm models the individual tree crown as a sphere or a hemisphere, which appears in a 2D circular form when projected onto a horizontal plane, and thus, the estimation problem is reduced to an issue of determining the diameter or radius of these projected circles. The following scheme is applied to achieve the goal of rough estimation of tree crown diameters (
Figure 8):
- 1.
Perform Delaunay triangulation of the 2D point set formed by estimated trunk positions within a partition and calculate each triangle’s center.
- 2.
Choose the tree trunk with the most LiDAR data points (indicating the highest degree of trueness) as the primary tree (#0 in
Figure 8a). Measure the distances from this tree location to centers of all triangles to which it belongs, and take the average of these distances as the radius or half of the diameter of the circle representing the tree’s crown region. In
Figure 8a,
is the estimate of the crown diameter for Tree #0.
- 3.
Use the primary tree as the root to generate a directed acyclic graph (DAG), as illustrated in
Figure 8b. Each vertex is assigned a specific rank, the value of which indicates its “distance” to the root. The symbol
appearing beside a directed edge connecting two vertices represents the mutual distance between two neighboring trees with indices of
i and
j.
- 4.
For the vertices with the rank value of 1, the crown diameters of their corresponding trees can be roughly determined as follows:
where
is the number of all vertices with a rank of 1.
- 5.
For other vertices, the rank values of which exceed 1, use the following formula to estimate their crown diameters:
where
is the total number of directed edges ending at Vertex #
k and
is the index of each directed edge’s starting vertex.
Next, an adaptive mean shift-based clustering analysis is performed on the dataset constituted by all separated tree crown points within a partition. A truncated, multivariate Gaussian function is adopted as the kernel or window in this stage. Each data point in the feature space will be associated with a specific kernel function, which is characterized by the adaptable kernel bandwidth (or window size), which can be varied according to the estimated tree crown diameter. Map the point onto the horizontal plane in
Figure 8a to see in which projected circular region it falls. If this point is not in any circle or belongs to two or more circles at the same time, assign the nearest circle to it. Assume the corresponding tree circle has the index number of
k, and then, determine the variable kernel bandwidth as follows:
where
is the variable kernel bandwidth. Please refer to [
26] for more detailed information about this adaptive mean shift algorithm.
An example of clustering results of the above-described adaptive mean shift algorithm is shown in
Figure 9. The major weakness of this clustering algorithm is that it tends to merge nearby trees or smaller trees into one single tree. Owing to the inability to detect suppressed trees or the difficulty in discriminating two overlapping tree crowns or oversized kernel bandwidths, omission errors (or under-segmentations) are very likely to happen in the clustering results (
Figure 10a,b). Besides, commission errors can also occur in the segmentation results of the adaptive mean shift-based clustering due to a loose canopy structure or because the derived kernel bandwidth somewhere is too small relative to its local canopy structure (
Figure 10c).
These problems originate from the spatial information contained in the dataset used by the algorithm, which mainly reflects the forest canopy’s structural characteristics. In order to tackle the weaknesses of the adaptive mean shift-based segmentation, additional information should be introduced to perform further analysis of LiDAR segments in the crown section. In this study, we utilize the tree trunk information to aid in the ultimate object-oriented processing, where the combinational use of trunk detection results and clustering results of crown points implies mutual reference based on which final decisions could be made to identify and delineate individual trees. Such a process aims to improve the overall detection accuracy, especially for structurally complex forests where most of the trees overlap. A detailed description of this technique is given in the following:
- 1.
Check the tree trunk detection results with respect to crown point segments. As the trunk area might not be reliably separated from the crown points and thus there could be errors in trunk detection, it makes sense to use the crown point cloud as a reference to verify the results of tree trunk detection. If there is no crown segment in the region right over an estimated tree trunk, which is defined as a circle with a radius of 0.5 m centered on the trunk’s approximate position, the trunk is considered as a dead tree or a low-growing plant and would be deleted from the trunk section. Any trunk cluster whose lowest point has an elevation value of more than
would be deemed to be a segment from the canopy and should be ignored. If a point cluster in the trunk section is composed of disjointed segments whose maximum elevation difference exceeds
(as depicted by the dashed line in
Figure 7b), we would regard it as a fictitious tree trunk.
- 2.
Link the point clusters in the crown section to the validated stem points in the trunk section. Firstly, determine the axis-aligned bounding box for each crown segment. Then, check to see if there are detected tree trunks in these bounding boxes. In case there is only one trunk in a bounding box, associate the trunk points with the crown points contained in the box and combine them to delineate the whole structure for an individual tree (
Figure 11a).
- 3.
If a segment in the crown section is not linked to any trunk points, it would be treated as a part of a tree crown. Merge this segment into the adjacent tree nearest to it. An example of point cloud merging is illustrated in
Figure 11b. According to this rule, the right segment rendered in green color, with no trunk under it, would be incorporated into a larger segment on the left.
- 4.
If there is more than one trunk contained in a bounding box, the point cloud in this box should be split into several parts, the exact number of which depends on the number of identified tree trunks. To achieve this goal, crown points are classified sequentially from bottom to top and will be allocated to the corresponding tree trunks. Each tree crown is built up concurrently beginning from its seed point, which is defined by the highest point of each identified trunk, by including nearby points and excluding points of other crowns based on their relative spacing. In the crown growing process, the spatial distances between the unclassified points and the semi-grown crown points are calculated. A point will be allocated to a target tree crown if the spatial distance to one of the points from that crown is the shortest. This minimum spacing rule-based tree crown growing process is illustrated in
Figure 11c.
3.7. Tree-Based Parameter Estimation
Once individual trees are accurately identified, tree structural attributes such as tree height, crown width and height and the number density of trees can be directly derived from the segmented points. From these parameters, other forest parameters, such as DBH, Crown Base Height (CBH), basal area, LAI, wood volume, biomass and species type, could be estimated from allometric equations. As our main objective was to develop a methodology to accurately segment individual trees from LiDAR data, we focus on two parameters for which we had in situ measured values from a ground survey, namely tree height and crown diameter.
The highest point among the segmented points for each tree is considered as the tree top, and the distance to the tree top from the ground is regarded as the height of that tree. Tree heights are commonly underestimated in LiDAR studies because the transmitted laser pulses may miss the actual tree top [
38]. Increasing the spatial density of LiDAR points provides a better chance of hitting the tree top, resulting in higher accuracy. The crown diameter is defined as the average of two values of crown widths measured along two perpendicular directions from the location of the tree trunk. The method to estimate individual tree crown diameter presented in [
7] is adopted in this study. Firstly, project the points from the topmost crown into the
x-
y plane, and extract perpendicular profiles of them centered on the tree trunk. Then, fit a fourth-degree polynomial on each profile, and find critical points of the fitted function around the center (namely the trunk position). Next, calculate crown width on each profile as the distance between critical points on each side of the center. Finally, the value for a crown diameter is computed as the average of the crown widths along the two directions or profile.
3.8. Implementation
A prototype system for 3D forest point cloud data processing was developed using the C/C++ programming language based on the Point Cloud Library (PCL) (Open Perception: Menlo Park, CA, USA), an open-source library of algorithms for point cloud processing tasks and 3D geometry processing, to verify our proposed approach, while the calibration and validation of the approach was performed in MATLAB (The MathWorks: Natick, MA, USA). Generic techniques for classification, filtering, segmentation, modeling and visualization of 3D point clouds were implemented with the methods and classes contained in PCL’s modules. The iterative computing procedures for the mean shift-based clustering were accomplished by calling a C++ wrapper that implements a standard mean shift algorithm. In particular, the approach-specific algorithms, such as tree trunk detection, kernel calibration and object-oriented post-processing, were implemented from scratch.
5. Discussion
This paper addresses the methods of airborne LiDAR-based remote sensing for individual tree-level forest inventory in which the detection of individual trees is one of the key issues. The mean shift-based clustering technique needs no seed points for initialization and works fully in three dimensions, and so, it is becoming a promising technique of producing relatively high accuracy by detecting more small trees in the lower and intermediate forest layer than other methods. Mean shift’s applications in individual tree detection using LiDAR data were explored in [
15,
24,
25]. In our previous work, we developed an adaptive mean shift-based clustering scheme, which automatically calibrated the kernel bandwidth to the local spatial structure of the forest canopy [
26]. The approach of tree trunk detection-aided mean shift clustering presented in this paper goes one step further than existing schemes by utilizing additional information below the tree crowns. To assess the overall performance of our proposed trunk-aided mean shift clustering approach, we compare the detection results of this approach with those obtained by several different existing approaches.
Firstly, the overall tree detection rates for trees located in different forest stories within six test subplots with IDs of DHS0101, DHS0102, DHS0201, DHS0202, DHS0301 and DHS0302 were compared between two mean shift-based clustering schemes (the proposed trunk-aided method in this study vs. the adaptive method presented in our previous study [
26]) to verify whether the trunk detection process works. As illustrated in
Table 3, although the detection rates of both methods decrease with dominance position, the trunk-aided method shows clearly higher accuracies than the other one for each forest story, especially for suppressed trees in the understory: nearly 13% more suppressed trees can be detected. This performance improvement is achieved by significantly reducing both the omission error (indicated by the FN count, reduced from 48 to 33) and commission errors (indicated by the FP count, reduced from 22 to 18). Furthermore, our proposed approach is compared to some representative approaches in terms of the accuracies of individual tree detection, as shown in
Table 4. In addition to the indicators of “recall” (
r) and “precision” (
p), the “F-score” (
) is used in the comparison to evaluate the overall detection accuracy. Except the adaptive mean shift-based clustering algorithm presented in [
26], the approaches chosen for this accuracy comparison include: a region growing approach presented in [
2], a
k-means clustering approach presented in [
19], a mean shift-based approach described in [
15] and a CHM-based approach adopted in [
40]. For more detailed descriptions about these individual tree detection approaches, please refer to [
26]. From the data listed in
Table 4, the proposed trunk-aided mean shift approach has the highest tree detection rates among all approaches used in this comparative experiment, and the accuracy improvement of our previous work (the adaptive mean shift-based clustering algorithm presented in [
26]) by applying the tree trunk detection strategy is noticeable: the overall detection accuracy in terms of “recall” is increased by 5%.
In order to compare the overall tree detection performance among different approaches in a statistically-rigorous fashion, the statistical significance of differences in detection accuracy was evaluated in terms of the overall accuracy metric: the “F-score”. As there is no parametric test suitable for this task, a Monte Carlo permutation test presented by [
41] was adopted here to determine the statistical significance of the difference between two F-scores. In this test, 999 permutations were generated, and for each permutation, the pairings of the actual trees with the predicted trees defined by two detection approaches were randomly shuffled; the F-scores were recomputed; and the difference between the two derived F-scores was estimated. Assuming the common situation in which a two-sided test (the null hypothesis that there is no significant difference between the two F-scores) at the five percent level of significance is undertaken, the difference between two F-scores derived with the same datasets would be regarded as being statistically significant if the computed proportion was less than 0.05. The results of a selection of pairwise comparisons are summarized in
Table 5.
Obviously, the proposed trunk-aided mean shift approach accounts for an accuracy improvement of the overall tree detection rate (see
Table 4 and
Table 5). Such an improvement of the tree detection performance could be expected since in many forest stands, neighboring trees with crowns overlapping each other do not appear as two distinct clusters in the point clouds, but their trunks can be clearly separated for the simple reason that the point structure of trunks is less ambiguous compared with crown shapes that are partially merged. The refinement of the detection rate is especially apparent for the intermediate and suppressed trees, as shown in
Table 3. Another strength of the proposed techniques is that the accuracy in tree positioning is significantly improved (see
Table 2). This is mainly due to the fact that the intersection of the detected tree trunk with the ground must be more precise than the approximate tree position derived from the stationary point of a data cluster.
The major limitation or the biggest uncertainty of the proposed approach is that not all tree trunks can be correctly detected. The performance of trunk extraction depends on the LiDAR point density and the forest type [
11,
28]. The trunk detection works successfully, if there are enough laser hits at the trunk and if the trunk area can be reliably separated from the crown points. Otherwise, when reflections of the laser beam in the trunk area are rare and trunk points cannot be clearly clustered, or when many suppressed or small trees are located below the taller trees, trunk detection would fail, and thus, the achieved improvement of the overall tree detection would be low. The second limitation, not just for this approach, but a common problem for individual tree-level forest inventory systems using aerial remote sensing technologies, is the under-segmentation issue [
27]: still, 8.9% of trees cannot be detected correctly in the studied forest plots using our approach. The reason is that laser beams are less likely to penetrate the forest canopy and hit suppressed trees, and this will disable the algorithms to extract the tree crowns or trunks in the segmentation stage. Other drawbacks of the approach are caused by the simplified removal of ground-covering vegetation, the simplified technique of determining the separating plane between the trunk and crown areas and insufficient rules to decide if a point group represents a trunk. Due to these drawbacks, a detected tree trunk may be fictitious, or the crown points belonging to a partition cannot be separated with respect to the detected trunks.
Performance evaluation of the tree-based structural parameter estimation can be conducted by statistics calculation and regression analysis. The linear fit for the tree height’s scatterplots reveals a slope nearly equal to one (1.0079) and an offset of 0.1379; this manifests a systematic underestimation of tree heights by the LiDAR data, which is consistent with other similar studies [
2,
15,
19,
40], as shown in
Table 6. The underestimates were due to the lower probability that an emitted laser pulse will strike the top of a tree rather than elsewhere on the tree crown. Additionally, each laser beam will penetrate tree foliage to some degree before being reflected back to the sensor. The regression analysis results also indicate that the average crown width is less accurately estimated, with a relative RMS error of 21.92%, than the tree height. Most of the variance associated with crown diameter can be attributed to the difference between overlapping and non-overlapping crown widths since LiDAR can only measure non-overlapping tree crowns (the algorithm for crown diameter estimation on the segmented LiDAR points aims at measuring the non-overlapping crowns), while the field measurements consider tree crowns to their full extent and therefore measured overlapping crown widths. Another portion of the variance for crown diameter can also be attributed to uncertainties in the ground surveys themselves because of random measurement errors induced by terrain conditions, measuring apparatus or field personnel. Compared with other approaches presented in the above-mentioned literature, the proposed trunk-aided mean shift approach could achieve higher accuracy in estimation of tree heights and crown diameters for individual trees (see
Table 6). This is mainly due to the more precise crown characterization ability offered by the proposed approach than other ones.
Further accuracy refinements for individual tree detection and tree parameter estimation could be achieved by increasing the average LiDAR point density, which refers to the nominal value influenced by the laser pulse repetition frequency, flight pattern, flying height, flying speed and strip overlap [
2,
11]. The common means to increase the LiDAR point density include reducing the flying speed, increasing the overlapping of laser strips, accelerating the emission of laser pulses, etc. Alternately, the overall point density could be increased by optimizing the flight pattern; for instance, flight lines can be surveyed twice or even more [
28]. Apparently, a laser scanner with a higher point density could provide sufficient data points reflected from the tree trunk and thereby optimize the trunk detection accuracy. Furthermore, a higher LiDAR point density increases the chance of a laser pulse return from the highest point in each tree canopy, thus reducing underestimation of tree heights. Moreover, further improvements of individual segmentation processes are possible by more sophisticated post-processing of the clustering results based on shape-related rules or using a priori knowledge about the forest. For instance, LiDAR clusters could be tested with respect to closeness. Furthermore, a cluster could not be accepted if its size exceeds tolerances about the tree shape. These issues will be addressed in our future research. Considering there are some limitations for tree segmentation based solely on LiDAR measurements, we also plan to use the proposed techniques in conjunction with aerial imagery, which provides better planform geometry and color (spectral) information, to further improve individual tree segmentation.