Extraction of Leaf Biophysical Attributes Based on a Computer Graphic-based Algorithm Using Terrestrial Laser Scanning Data
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Site and Data Collection
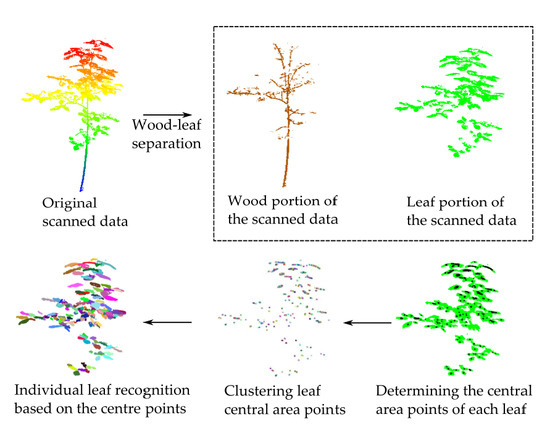
2.2. Wood-leaf Separation
2.3. Individual Leaf Segmentation
2.3.1. Extracting the Central Area Points of Individual Leaves
2.3.2. Clustering of Leaf Central Area Points
2.3.3. Individual Leaf Segmentation
2.4. Leaf Attributes Calculation Based on Classified Leaf Points
3. Results
3.1. Plant Leaf Classification
3.2. Leaf Segmentation
3.3. Leaf Attribute Calculation
4. Discussion
4.1. Tree Species Effects
4.2. Sensitivity Analysis of Search Radius ()
4.3. Recommendations
5. Conclusions
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Han, W.T.; Sun, Y.; Xu, T.F.; Chen, X.W.; Su, K.O. Detecting maize leaf water status by using digital RGB images. Int. J. Agric. Biol. Eng. 2014, 7, 45–53. [Google Scholar] [CrossRef]
- Harley, P.C.; Baldocchi, D.D. Scaling carbon dioxide and water vapour exchange from leaf to canopy in a deciduous forest. I. Leaf model parametrization. Plant Cell Environ. 2010, 18, 1146–1156. [Google Scholar] [CrossRef]
- Huang, J.; Sedano, F.; Huang, Y.; Ma, H.; Li, X.; Liang, S.; Tian, L.; Zhang, X.; Fan, J.; Wu, W. Assimilating a synthetic Kalman filter leaf area index series into the WOFOST model to improve regional winter wheat yield estimation. Agric. For. Meteorol. 2016, 216, 188–202. [Google Scholar] [CrossRef]
- Strebel, D.E.; Goel, N.S.; Ranson, K.J. Two-Dimensional Leaf Orientation Distributions. IEEE Trans. Geosci. Remote Sens. 1985, 23, 640–647. [Google Scholar] [CrossRef]
- Ross, J. The Radiation Regime and Architecture of Plant Stands; Springer: Cham, The Netherlands, 1981. [Google Scholar] [CrossRef]
- Lugg, D.G.; Youngman, V.E.; Hinze, G. Leaf Azimuthal Orientation of Sorghum in Four Row Directions. Agron. J. 1981, 73, 497–500. [Google Scholar] [CrossRef]
- Chianucci, F.; Cutini, A.; Corona, P.; Puletti, N. Estimation of leaf area index in understory deciduous trees using digital photography. Agric. For. Meteorol. 2014, 198–199, 259–264. [Google Scholar] [CrossRef]
- Peper, P.J.; Mcpherson, E.G. Evaluation of four methods for estimating leaf area of isolated trees. Urban For. Urban Green. 2004, 2, 19–29. [Google Scholar] [CrossRef]
- Colaizzi, P.D.; Evett, S.R.; Brauer, D.K.; Howell, T.A.; Tolk, J.A.; Copeland, K.S. Allometric Method to Estimate Leaf Area Index for Row Crops. Agron. J. 2017, 109. [Google Scholar] [CrossRef]
- Xie, D.; Wang, Y.; Hu, R.; Chen, Y.; Yan, G.; Zhang, W.; Wang, P. Modified gap fraction model of individual trees for estimating leaf area using terrestrial laser scanner. J. Appl. Remote Sens. 2017, 11. [Google Scholar] [CrossRef]
- Macfarlane, C.; Ryu, Y.; Ogden, G.N.; Sonnentag, O. Digital canopy photography: Exposed and in the raw. Agric. For. Meteorol. 2014, 197, 244–253. [Google Scholar] [CrossRef]
- Stenberg, P. Correcting LAI-2000 estimates for the clumping of needles in shoots of conifers. Agric. For. Meteorol. 1996, 79, 1–8. [Google Scholar] [CrossRef]
- Chen, J.M.; Cihlar, J. Quantifying the effect of canopy architecture on optical measurements of leaf area index using two gap size analysis methods. IEEE Trans. Geosci. Remote Sens. 1995, 33, 777–787. [Google Scholar] [CrossRef]
- Yun, T.; Li, W.; Sun, Y.; Xue, L. Study of Subtropical Forestry Index Retrieval Using Terrestrial Laser Scanning and Hemispherical Photography. Math. Probl. Eng. 2015, 2015, 206108. [Google Scholar] [CrossRef]
- Mason, E.G.; Diepstraten, M.; Pinjuv, G.L.; Lasserre, J.P. Comparison of direct and indirect leaf area index measurements of Pinus radiata D. Don. Agric. For. Meteorol. 2012, 166–167, 113–119. [Google Scholar] [CrossRef]
- Asner, G.P.; Scurlock, J.M.O.; Hicke, J.A. Global synthesis of leaf area index observations: Implications for ecological and remote sensing studies. Glob. Ecol. Biogeogr. 2010, 12, 191–205. [Google Scholar] [CrossRef]
- Zhao, K.; García, M.; Liu, S.; Guo, Q.; Chen, G.; Zhang, X.; Zhou, Y.; Meng, X. Terrestrial lidar remote sensing of forests: Maximum likelihood estimates of canopy profile, leaf area index, and leaf angle distribution. Agric. For. Meteorol. 2015, 209–210, 100–113. [Google Scholar] [CrossRef]
- Ali, A.M.; Darvishzadeh, R.; Skidmore, A.K.; van Duren, I. Specific leaf area estimation from leaf and canopy reflectance through optimization and validation of vegetation indices. Agric. For. Meteorol. 2017, 236, 162–174. [Google Scholar] [CrossRef]
- Norman, J.M.; Campbell, G.S. Canopy Structure-in Plant Physiological Ecology: Field Methods Instrumentation; Chapman and Hall: London, UK, 1989; pp. 301–325. [Google Scholar]
- Lang, A.R.G. Leaf orientation of a cotton plant. Agric. Meteorol. 1973, 11, 37–51. [Google Scholar] [CrossRef]
- Zou, X.; Mõttus, M. Retrieving crop leaf tilt angle from imaging spectroscopy data. Agric. For. Meteorol. 2015, 205, 73–82. [Google Scholar] [CrossRef]
- Pisek, J.; Ryu, Y.; Alikas, K. Estimating leaf inclination and G-function from leveled digital camera photography in broadleaf canopies. Trees 2011, 25, 919–924. [Google Scholar] [CrossRef]
- Raabe, K.; Pisek, J.; Sonnentag, O.; Annuk, K. Variations of leaf inclination angle distribution with height over the growing season and light exposure for eight broadleaf tree species. Agric. For. Meteorol. 2015, 214–215, 2–11. [Google Scholar] [CrossRef]
- Daughtry, C.S.T. Direct measurements of canopy structure. Remote Sens. Rev. 1990, 5, 45–60. [Google Scholar] [CrossRef]
- Zheng, G.; Moskal, L.M.; Kim, S.H. Retrieval of Effective Leaf Area Index in Heterogeneous Forests With Terrestrial Laser Scanning. IEEE Trans. Geosci. Remote Sens. 2013, 51, 777–786. [Google Scholar] [CrossRef]
- Bayer, D.; Seifert, S.; Pretzsch, H. Structural crown properties of Norway spruce (Picea abies L. Karst.); and European beech (Fagus sylvatica L.) in mixed versus pure stands;revealed by terrestrial laser scanning. Trees 2013, 27, 1035–1047. [Google Scholar] [CrossRef]
- Wezyk, P.; Koziol, K.; Glista, M.; Pierzchalski, M. Terrestrial laser scanning versus traditional forest inventory first results from the polish forests. In Proceedings of the ISPRS Workshop on Laser Scanning, 12–14 September 2007, Espoo, Finland.
- Liu, G.; Wang, J.; Dong, P.; Chen, Y.; Liu, Z. Estimating Individual Tree Height and Diameter at Breast Height (DBH) from Terrestrial Laser Scanning (TLS) Data at Plot Level. Forests 2018, 9, 398. [Google Scholar] [CrossRef]
- Zheng, G.; Moskal, L.M. Retrieving Leaf Area Index (LAI) Using Remote Sensing: Theories, Methods and Sensors. Sensors 2009, 9, 2719–2745. [Google Scholar] [CrossRef] [Green Version]
- Hosoi, F.; Nakai, Y.; Omasa, K. Estimating the leaf inclination angle distribution of the wheat canopy using a portable scanning lidar. Agric. Meteorol. 2009, 65, 297–302. [Google Scholar] [CrossRef] [Green Version]
- Janowski, A.; Bobkowska, K.; Szulwic, J. 3D modelling of cylindrical-shaped objects from lidar data—An assessment based on theoretical modelling and experimental data. Metrol. Meas. Syst. 2018, 25, 47–56. [Google Scholar]
- Janowski, A. The circle object detection with the use of Msplit estimation. E3S Web Conf. 2018, 26, 00014. [Google Scholar] [CrossRef] [Green Version]
- Liang, X.; Kankare, V.; Hyyppä, J.; Wang, Y.; Kukko, A.; Haggrén, H.; Yu, X.; Kaartinen, H.; Jaakkola, A.; Guan, F.; et al. Terrestrial laser scanning in forest inventories. ISPRS J. Photogramm. Remote Sens. 2016, 115, 63–77. [Google Scholar] [CrossRef] [Green Version]
- Bailey, B.N.; Mahaffee, W.F. Rapid measurement of the three-dimensional distribution of leaf orientation and the leaf angle probability density function using terrestrial LiDAR scanning. Remote Sens. Environ. 2017, 194, 63–76. [Google Scholar] [CrossRef]
- Sala, F.; Arsene, G.G.; Iordănescu, O.; Boldea, M. Leaf area constant model in optimizing foliar area measurement in plants A case study in apple tree. Sci. Hortic. 2015, 193, 218–224. [Google Scholar] [CrossRef]
- Keramatlou, I.; Sharifani, M.; Sabouri, H.; Alizadeh, M.; Kamkar, B. A simple linear model for leaf area estimation in Persian walnut (Juglans regia L.). Sci. Hortic. 2015, 184, 36–39. [Google Scholar] [CrossRef]
- Pompelli, M.F.; Antunes, W.C.; Ferreira, D.T.R.G.; Cavalcante, P.G.S.; Wanderley-Filho, H.C.L.; Endres, L. Allometric models for non-destructive leaf area estimation of Jatropha curcas. Biomass Bioenergy 2012, 36, 77–85. [Google Scholar] [CrossRef]
- Roberts, S.D.; Dean, T.J.; Evans, D.L. Family influences on leaf area estimates derived from crown and tree dimensions in Pinus taeda. For. Ecol. Manag. 2003, 172, 261–270. [Google Scholar] [CrossRef]
- Liang, W.Z.; Kirk, K.R.; Greene, J.K. Estimation of soybean leaf area, edge, and defoliation using color image analysis. Comput. Electron. Agric. 2018, 150, 41–51. [Google Scholar] [CrossRef]
- Béland, M.; Widlowski, J.L.; Fournier, R.A.; Côté, J.F.; Verstraete, M.M. Estimating leaf area distribution in savanna trees from terrestrial LiDAR measurements. Agric. For. Meteorol. 2011, 151, 1252–1266. [Google Scholar] [CrossRef]
- Béland, M.; Baldocchi, D.D.; Widlowski, J.L.; Fournier, R.A.; Verstraete, M.M. On seeing the wood from the leaves and the role of voxel size in determining leaf area distribution of forests with terrestrial LiDAR. Agric. For. Meteorol. 2014, 184, 82–97. [Google Scholar] [CrossRef]
- Ma, L.; Zheng, G.; Wang, X.; Li, S.; Lin, Y.; Ju, W. Retrieving forest canopy clumping index using terrestrial laser scanning data. Remote Sens. Environ. 2018, 210, 452–472. [Google Scholar] [CrossRef]
- Sanz, R.; Llorens, J.; Escolà, A.; Arnó, J.; Planas, S.; Román, C.; Rosell-Polo, J.R. LIDAR and non-LIDAR-based canopy parameters to estimate the leaf area in fruit trees and vineyard. Agric. For. Meteorol. 2018, 260–261, 229–239. [Google Scholar] [CrossRef]
- Kishor, K.M.; Senthil, K.R.; Sankar, V.; Sakthivel, T.; Karunakaran, G.; Tripathi, P.C. Non-destructive estimation of leaf area of durian (Durio zibethinus)—An artificial neural network approach. Sci. Hortic. 2017, 219, 319–325. [Google Scholar] [CrossRef]
- Shabani, A.; Ghaffary, K.A.; Sepaskhah, A.R.; Kamgar-Haghighi, A.A. Using the artificial neural network to estimate leaf area. Sci. Hortic. 2017, 216, 103–110. [Google Scholar] [CrossRef]
- Huang, W.; Niu, Z.; Wang, J.; Liu, L.; Zhao, C.; Liu, Q. Identifying Crop Leaf Angle Distribution Based on Two-Temporal and Bidirectional Canopy Reflectance. IEEE Trans. Geosci. Remote Sens. 2006, 44, 3601–3609. [Google Scholar] [CrossRef]
- Bacour, C.; Jacquemoud, S.; Leroy, M.; Hautecœur, O.; Weiss, M.; Prévot, L.; Bruguier, N.; Chauki, H. Reliability of the estimation of vegetation characteristics by inversion of three canopy reflectance models on airborne POLDER data. Agronomie 2002, 22, 555–566. [Google Scholar] [CrossRef]
- Lang, A. Leaf area and average leaf angle from transmission of direct sunlight. Aust. J. Bot. 1986, 34, 349–355. [Google Scholar] [CrossRef]
- Ryu, Y.R.; Sonnentag, O.; Nilson, T.; Vargas, R.; Kobayashi, H.; Wenk, R.; Baldocchi, D.D. How to quantify tree leaf area index in an open savanna ecosystem: A multi-instrument and multi-model approach. Agric. For. Meteorol. 2010, 150, 63–76. [Google Scholar] [CrossRef]
- Zheng, G.; Moskal, L.M. Leaf Orientation Retrieval From Terrestrial Laser Scanning (TLS) Data. IEEE Trans. Geosci. Remote Sens. 2012, 50, 3970–3979. [Google Scholar] [CrossRef]
- Hosoi, F.; Omasa, K. Estimating leaf inclination angle distribution of broad-leaved trees in each part of the canopies by a high-resolution portable scanning lidar. J. Agric. Meteorol. 2015, 71, 136–141. [Google Scholar] [CrossRef] [Green Version]
- Tao, S.; Guo, Q.; Su, Y.; Xu, S.; Li, Y.; Wu, F. A Geometric Method for Wood-Leaf Separation Using Terrestrial and Simulated Lidar Data. Photogramm. Eng. Remote Sens. 2015, 81, 767–776. [Google Scholar] [CrossRef]
- Yun, T.; An, F.; Li, W.; Sun, Y.; Cao, L.; Xue, L. A Novel Approach for Retrieving Tree Leaf Area from Ground-Based LiDAR. Remote Sens. 2016, 8, 942. [Google Scholar] [CrossRef]
- Piayda, A.; Dubbert, M.; Werner, C.; Correia, A.V.; Pereira, J.S.; Cuntz, M. Influence of woody tissue and leaf clumping on vertically resolved leaf area index and angular gap probability estimates. For. Ecol. Manag. 2015, 340, 103–113. [Google Scholar] [CrossRef]
- Woodgate, W.; Disney, M.; Armston, J.D.; Jones, S.D.; Suarez, L.; Hill, M.J.; Mellor, A. An improved theoretical model of canopy gap probability for Leaf Area Index estimation in woody ecosystems. For. Ecol. Manag. 2015, 358, 303–320. [Google Scholar] [CrossRef]
- Ester, M.; Kriegel, H.P.; Xu, X. A Density-Based Algorithm for Discovering Clusters in Large Spatial Databases with Noise. In Proceedings of the International Conference on Knowledge Discovery and Data Mining, Portland, OR, USA, 2–4 August 1996; pp. 226–231. [Google Scholar]
- Daszykowski, M.; Walczak, B.; L, D. Massart Looking for Natural Patterns in Analytical Data. 2. Tracing Local Density with OPTICS. J. Chem. Inf. Comput. Sci. 2016, 42, 500. [Google Scholar] [CrossRef]
- Quesada-Barriuso, P.; Heras, D.B. Efficient 2D and 3D Watershed on Graphics Processing Unit: Block-Asynchronous Approaches Based on Cellular Automata; Pergamon Press Inc.: New York, NY, USA, 2013. [Google Scholar] [CrossRef]
- Moumoun, R.; El Far, L.; Chahhou, M.; Gadi, M.; Benslimane, T. Solving the 3D watershed over-segmentation problem using the generic adjacency graph. In Proceedings of the 2010 5th International Symposium On I/V Communications and Mobile Network, Rabat, Morocco, 30 September–2 October 2010. [Google Scholar] [CrossRef]
- Lin, W.; Meng, Y.; Qiu, Z.; Zhang, S.; Wu, J. Measurement and calculation of crown projection area and crown volume of individual trees based on 3D laser-scanned point-cloud data. Int. J. Remote Sens. 2017, 38, 1083–1100. [Google Scholar] [CrossRef]
- Fiorani, F.; Schurr, U. Future scenarios for plant phenotyping. Annu. Rev. Plant Biol. 2013, 64, 267. [Google Scholar] [CrossRef] [PubMed]
- Fiorani, F.; Rascher, U.; Jahnke, S.; Schurr, U. Imaging plants dynamics in heterogenic environments. Curr. Opin. Biotechnol. 2012, 23, 227–235. [Google Scholar] [CrossRef]
- Houborg, R.; Soegaard, H.; Boegh, E. Combining vegetation index and model inversion methods for the extraction of key vegetation biophysical parameters using Terra and Aqua MODIS reflectance data. Remote Sens. Environ. 2007, 106, 39–58. [Google Scholar] [CrossRef]
- Asner, G.P. Biophysical and Biochemical Sources of Variability in Canopy Reflectance. Remote Sens. Environ. 1998, 64, 234–253. [Google Scholar] [CrossRef]
- Fanourakis, D.; Heuvelink, E.; Carvalho, S.M.P. Spatial heterogeneity in stomatal features during leaf elongation: An analysis using Rosa hybrida. Funct. Plant Biol. 2015, 42, 737–745. [Google Scholar] [CrossRef]
- Fanourakis, D.; Bouranis, D.; Giday, H.; Carvalho, D.R.; Rezaei, N.A.; Ottosen, C.O. Improving stomatal functioning at elevated growth air humidity: A review. J. Plant Physiol. 2016, 207, 51–60. [Google Scholar] [CrossRef]
- De Oliveira Silva, F.M.; Lichtenstein, G.; Alseekh, S.; Rosado-Souza, L.; Conte, M.; Suguiyama, V.F.; Lira, B.S.; Fanourakis, D.; Usadel, B.; Bhering, L.L.; et al. The Genetic Architecture of Photosynthesis and Plant Growth Related Traits in Tomato. Plant Cell Environ. 2017, 41, 327–341. [Google Scholar] [CrossRef] [PubMed]
- Danson, F.M.; Hetherington, D.; Morsdorf, F.; Koetz, B.; Allgower, B. Forest Canopy Gap Fraction From Terrestrial Laser Scanning. IEEE Geosci. Remote Sens. Lett. 2007, 4, 157–160. [Google Scholar] [CrossRef] [Green Version]
- Bremer, M.; Wichmann, V.; Rutzinger, M. Calibration and Validation of a Detailed Architectural Canopy Model Reconstruction for the Simulation of Synthetic Hemispherical Images and Airborne LiDAR Data. Remote Sens. 2017, 9, 220. [Google Scholar] [CrossRef]
- Watt, P.J. Measuring forest structure with terrestrial laser scanning. Int. J. Remote Sens. 2005, 26, 1437–1446. [Google Scholar] [CrossRef]
- Kankare, V.; Holopainen, M.; Vastaranta, M.; Puttonen, E.; Yu, X.; Hyyppä, J.; Vaaja, M.; Hyyppä, H.; Alho, P. Individual tree biomass estimation using terrestrial laser scanning. ISPRS J. Photogramm. Remote Sens. 2013, 75, 64–75. [Google Scholar] [CrossRef]
- Ma, L.; Zheng, G.; Eitel, J.U.H.; Moskal, L.M.; He, W.; Huang, H. Improved Salient Feature-Based Approach for Automatically Separating Photosynthetic and Nonphotosynthetic Components Within Terrestrial Lidar Point Cloud Data of Forest Canopies. IEEE Trans. Geosci. Remote Sens. 2016, 54, 679–696. [Google Scholar] [CrossRef]
- Cao, L.; Gao, S.; Li, P.; Yun, T.; Shen, X.; Ruan, H. Aboveground Biomass Estimation of Individual Trees in a Coastal Planted Forest Using Full-Waveform Airborne Laser Scanning Data. Remote Sens. 2016, 8, 729. [Google Scholar] [CrossRef]
- Ferrara, R.; Virdis, S.G.P.; Ventura, A.; Ghisu, T.; Duce, P.; Pellizzaro, G. An automated approach for wood-leaf separation from terrestrial LIDAR point clouds using the density based clustering algorithm DBSCAN. Agric. For. Meteorol. 2018. [Google Scholar] [CrossRef]
- Putmana, G.W.M.E.B.; Popescub, S.C.; Erikssonc, M.; Zhoua, T.; Klockowd, P.; Vogele, J. Detecting and quantifying standing dead tree structural loss with reconstructed tree models using voxelized terrestrial lidar data. Remote Sens. Environ. 2018, 209, 52–65. [Google Scholar] [CrossRef]









| Tree Species | Ehretia Macrophylla | Crape Myrtle | Fatsia Japonica |
|---|---|---|---|
| Total number of points | 36,604 | 107,914 | 247,133 |
| Tree height (m) | 3.5 | 3.42 | 1.6 |
| Crown projection area (m2) | 1.274 | 1.975 | 3.126 |
| Average spatial sampling (cm) | 0.55 | 0.45 | 0.42 |
| Leaf point number (classified/actual) | 27,841/29,387 | 72,639/78,564 | 184,721/201,648 |
| Branch point number (classified/actual) | 8763/7217 | 35,275/29,350 | 62,412/45,485 |
| Overall accuracy | 94.73% | 92.46% | 91.61% |
| Tree Species | Ehretia Macrophylla | Crape Myrtle | Fatsia Japonica | |||
|---|---|---|---|---|---|---|
| Upper | Middle | Lower | Upper | Lower | Tree Crown | |
| NP | 12,477 | 14,316 | 3438 | 35,028 | 37,639 | 103,177 |
| PD (pts·m−2) | 4320 | 6260 | 6531 | 5844 | 9410 | 51,484 |
| NLUM/ NLMM | 44/ 43 | 87/ 96 | 27/ 29 | 501/ 543 | 440/ 495 | 187/ 166 |
| RELN | 2.33% | 9.38% | 6.90% | 7.73% | 11.11% | 12.65% |
| RELL | 5.57% | 3.48% | 1.16% | 1.75% | 15.1% | 1.44% |
| RELW | 2.75% | 10.85% | 3.55% | 0.55% | 0.93% | 0.93% |
| RELA | 2.67% | 6.98% | 4.67% | 2.26% | 16.12% | 7.7% |
| LIAUM/ LIAMM (°) | 28.4–88.0/ 25.1–86.8 | 9.1–89.8/ 7.5–89.0 | 9.3–81.2/ 2.7–77.8 | 7.9–89.7/ 11.7–88.2 | 5.2–84.0/ 1.2–89.8 | 9.6–87.8/ 6.8–89.2 |
| LAAUM/ LAAMM (°) | 44.8–322.0/ 31.3–313.5 | 11.8–349.6/ 3.3–353.6 | 14.7–324.5/ 15.6–323.8 | 18.6–227.0/ 12.7–269.4 | 19.7–295.7/ 23.0–309.3 | 53.3–268.8/ 50.9–266.3 |
| Tree species | Position | R2 | RMSE | ||||
|---|---|---|---|---|---|---|---|
| LA | LIA | LAA | LA (cm2) | LIA (°) | LAA (°) | ||
| Ehretia macrophylla | Upper | 0.987 | 0.894 | 0.990 | 8.805 | 5.686 | 7.709 |
| Middle | 0.959 | 0.914 | 0.978 | 7.596 | 6.489 | 7.316 | |
| Lower | 0.888 | 0.831 | 0.982 | 9.357 | 7.841 | 8.151 | |
| All | 0.971 | 0.908 | 0.981 | 8.508 | 6.806 | 7.680 | |
| Crape myrtle | Upper | 0.934 | 0.864 | 0.941 | 7.042 | 8.459 | 7.011 |
| Lower | 0.801 | 0.905 | 0.934 | 4.579 | 8.260 | 8.180 | |
| All | 0.873 | 0.901 | 0.938 | 6.001 | 8.365 | 7.573 | |
| Fatsia japonica | All | 0.899 | 0.849 | 0.947 | 5.744 | 6.158 | 3.946 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, Q.; Cao, L.; Xue, L.; Chen, B.; An, F.; Yun, T. Extraction of Leaf Biophysical Attributes Based on a Computer Graphic-based Algorithm Using Terrestrial Laser Scanning Data. Remote Sens. 2019, 11, 15. https://0-doi-org.brum.beds.ac.uk/10.3390/rs11010015
Xu Q, Cao L, Xue L, Chen B, An F, Yun T. Extraction of Leaf Biophysical Attributes Based on a Computer Graphic-based Algorithm Using Terrestrial Laser Scanning Data. Remote Sensing. 2019; 11(1):15. https://0-doi-org.brum.beds.ac.uk/10.3390/rs11010015
Chicago/Turabian StyleXu, Qiangfa, Lin Cao, Lianfeng Xue, Bangqian Chen, Feng An, and Ting Yun. 2019. "Extraction of Leaf Biophysical Attributes Based on a Computer Graphic-based Algorithm Using Terrestrial Laser Scanning Data" Remote Sensing 11, no. 1: 15. https://0-doi-org.brum.beds.ac.uk/10.3390/rs11010015