Extending ALS-Based Mapping of Forest Attributes with Medium Resolution Satellite and Environmental Data
Abstract
:1. Introduction
2. Materials and Methods
2.1. Geographic Area
2.2. Ground Plots
2.3. ALS Data
2.4. Spatially Comprehensive Data
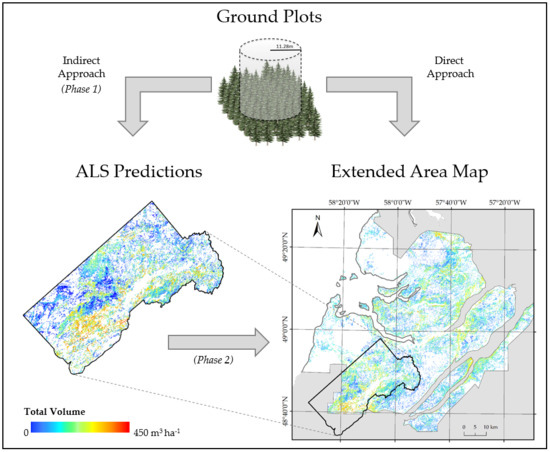
2.5. Overview of the Approach
2.6. Development of ALS-Based Inventory (Phase 1)
2.7. Development of Extended Inventory (Phase 2)
2.8. Independent Plot Evaluation
3. Results
3.1. ALS Models of Forest Attributes (Phase 1)
3.2. Satellite and Environmental Models (Phase 2)
3.3. Direct vs. Indirect Approach
3.4. Landscape Patterns
4. Discussion
4.1. ALS-Based Inventory
4.2. Extension of ALS-Based Inventory
4.3. Parametric vs. Nonparametric Models
4.4. Data and Technical Considerations
4.5. Implications for Forest Inventory
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Naesset, E. Airborne laser scanning as a method in operational forest inventory: Status of accuracy assessments accomplished in Scandinavia. Scand. J. For. Res. 2007, 22, 433–442. [Google Scholar] [CrossRef]
- Wulder, M.A.; Bater, C.W.; Coops, N.C.; Hilker, T.; White, J.C. The role of LiDAR in sustainable forest management. For. Chron. 2008, 84, 807–826. [Google Scholar] [CrossRef] [Green Version]
- Montaghi, A.; Corona, P.; Dalponte, M.; Gianelle, D.; Chirici, G.; Olsson, H. Airborne laser scanning of forest resources: An overview of research in Italy as a commentary case study. Int. J. Appl. Earth Obs. Geoinf. 2013, 23, 288–300. [Google Scholar] [CrossRef] [Green Version]
- Maltamo, M.; Næsset, E.; Vauhkonen, J. Forestry Applications of Airborne Laser Scanning: Concepts and Case Studies; Springer: Dordrecht, The Netherlands, 2014; Volume 27, ISBN 9789401786621. [Google Scholar]
- Næsset, E.; Gobakken, T. Estimation of above- and below-ground biomass across regions of the boreal forest zone using airborne laser. Remote Sens. Environ. 2008, 112, 3079–3090. [Google Scholar] [CrossRef]
- Woods, M.; Pitt, D.; Penner, M.; Lim, K.; Nesbitt, D.; Etheridge, D.; Treitz, P. Operational Implementation of a LiDAR Inventory in Boreal Ontario. For. Chron. 2011, 87, 512–528. [Google Scholar] [CrossRef]
- Luther, J.E.; Skinner, R.; Fournier, R.A.; van Lier, O.R.; Bowers, W.W.; Coté, J.F.; Hopkinson, C.; Moulton, T. Predicting wood quantity and quality attributes of balsam fir and black spruce using airborne laser scanner data. Forestry 2014, 87, 313–326. [Google Scholar] [CrossRef]
- Næsset, E. Predicting forest stand characteristics with airborne scanning laser using a practical two-stage procedure and field data. Remote Sens. Environ. 2002, 80, 88–99. [Google Scholar] [CrossRef]
- Holmgren, J. Prediction of tree height, basal area and stem volume in forest stands using airborne laser scanning. Scand. J. For. Res. 2004, 19, 543–553. [Google Scholar] [CrossRef]
- Hyyppä, J.; Hyyppä, H.; Leckie, D.; Gougeon, F.; Yu, X.; Maltamo, M. Review of methods of small-footprint airborne laser scanning for extracting forest inventory data in boreal forests. Int. J. Remote Sens. 2008, 29, 1339–1366. [Google Scholar] [CrossRef]
- White, J.C.; Wulder, M.A.; Varhola, A.; Vastaranta, M.; Coops, N.C.; Cook, B.D.; Pitt, D.; Woods, M. A Best Practices Guide for Generating Forest Inventory Attributes from Airborne Laser Scanning Data Using an Area-Based Approach; Information Report FI-X-010; Natural Resources Canada, Canadian Forest Service, Canadian Wood Fibre Centre: Victoria, BC, Canada, 2013; p. 41. [Google Scholar]
- Bouvier, M.; Durrieu, S.; Fournier, R.A.; Renaud, J.P. Generalizing predictive models of forest inventory attributes using an area-based approach with airborne LiDAR data. Remote Sens. Environ. 2015, 156, 322–334. [Google Scholar] [CrossRef]
- Li, Y.; Andersen, H.-E.; McGaughey, R. A comparison of statistical methods for estimating forest biomass from light detection and ranging data. J. Appl. For. 2008, 23, 223–231. [Google Scholar]
- Brosofske, K.D.; Froese, R.E.; Falkowski, M.J.; Banskota, A. A review of methods for mapping and prediction of inventory attributes for operational forest management. For. Sci. 2014, 60, 733–756. [Google Scholar] [CrossRef]
- Hansen, E.H.; Ene, L.T.; Mauya, E.W.; Patočka, Z.; Mikita, T.; Gobakken, T.; Næsset, E. Comparing empirical and semi-empirical approaches to forest biomass modelling in different biomes using airborne laser scanner data. Forests 2017, 8, 1–17. [Google Scholar] [CrossRef]
- Hollaus, M.; Dorigo, W.; Wagner, W.; Schadauer, K.; Höfle, B.; Maier, B. Operational wide-area stem volume estimation based on airborne laser scanning and national forest inventory data. Int. J. Remote Sens. 2009, 30, 5159–5175. [Google Scholar] [CrossRef]
- White, J.C.; Coops, N.C.; Wulder, M.A.; Vastaranta, M.; Hilker, T.; Tompalski, P. Remote Sensing Technologies for Enhancing Forest Inventories: A Review. Can. J. Remote Sens. 2016, 42, 619–641. [Google Scholar] [CrossRef] [Green Version]
- Kangas, A.; Astrup, R.; Breidenbach, J.; Fridman, J.; Gobakken, T.; Korhonen, K.T.; Maltamo, M.; Nilsson, M.; Nord-Larsen, T.; Næsset, E.; et al. Remote sensing and forest inventories in Nordic countries–roadmap for the future. Scand. J. For. Res. 2018, 33, 397–412. [Google Scholar] [CrossRef]
- Hudak, A.T.; Lefsky, M.A.; Cohen, W.B.; Berterretche, M. Integration of lidar and Landsat ETM+ data for estimating and mapping forest canopy height. Remote Sens. Environ. 2002, 82, 397–416. [Google Scholar] [CrossRef] [Green Version]
- Hudak, A.T.; Crookston, N.L.; Evans, J.S.; Falkowski, M.J.; Smith, A.M.S.; Gessler, P.E.; Morgan, P. Regression modeling and mapping of coniferous forest basal area and tree density from discrete-return lidar and multispectral satellite data. Can. J. Remote Sens. 2006, 32, 126–138. [Google Scholar] [CrossRef]
- Huang, S.; Ramirez, C.; Kennedy, K.; Mallory, J. A New Approach to Extrapolate Forest Attributes from Field Inventory with Satellite and Auxiliary Data Sets. For. Sci. 2017, 63, 232–240. [Google Scholar] [CrossRef]
- Zheng, D.; Rademacher, J.; Chen, J.; Crow, T.; Bresee, M.; Le Moine, J.; Ryu, S.R. Estimating aboveground biomass using Landsat 7 ETM+ data across a managed landscape in northern Wisconsin, USA. Remote Sens. Environ. 2004, 93, 402–411. [Google Scholar] [CrossRef]
- Labrecque, S.; Fournier, R.A.; Luther, J.E.; Piercey, D. A comparison of four methods to map biomass from Landsat-TM and inventory data in western Newfoundland. For. Ecol. Manage. 2006, 226, 129–144. [Google Scholar] [CrossRef]
- Lu, D.; Chen, Q.; Wang, G.; Moran, E.; Batistella, M.; Zhang, M.; Vaglio Laurin, G.; Saah, D. Aboveground forest biomass estimation with Landsat and LiDAR data and uncertainty analysis of the estimates. Int. J. For. Res. 2012, 2012, 1–16. [Google Scholar] [CrossRef]
- McRoberts, R.E. Estimating forest attribute parameters for small areas using nearest neighbors techniques. For. Ecol. Manage. 2012, 272, 3–12. [Google Scholar] [CrossRef]
- Beaudoin, A.; Bernier, P.Y.; Guindon, L.; Villemaire, P.; Guo, X.J.; Stinson, G.; Bergeron, T.; Magnussen, S.; Hall, R.J. Mapping attributes of Canada’s forests at moderate resolution through kNN and MODIS imagery. Can. J. For. Res. 2014, 44, 521–532. [Google Scholar] [CrossRef]
- Chirici, G.; Mura, M.; McInerney, D.; Py, N.; Tomppo, E.O.; Waser, L.T.; Travaglini, D.; McRoberts, R.E. A meta-analysis and review of the literature on the k-Nearest Neighbors technique for forestry applications that use remotely sensed data. Remote Sens. Environ. 2016, 176, 282–294. [Google Scholar] [CrossRef]
- Latifi, H.; Nothdurft, A.; Koch, B. Non-parametric prediction and mapping of standing timber volume and biomass in a temperate forest: Application of multiple optical/LiDAR-derived predictors. Forestry 2010, 83, 395–407. [Google Scholar] [CrossRef]
- Mäkelä, H.; Pekkarinen, A. Estimation of forest stand volumes by Landsat TM imagery and stand-level field-inventory data. For. Ecol. Manage. 2004, 196, 245–255. [Google Scholar] [CrossRef]
- Tomppo, E.; Olsson, H.; Ståhl, G.; Nilsson, M.; Hagner, O.; Katila, M. Combining national forest inventory field plots and remote sensing data for forest databases. Remote Sens. Environ. 2008, 112, 1982–1999. [Google Scholar] [CrossRef]
- Mura, M.; Bottalico, F.; Giannetti, F.; Bertani, R.; Giannini, R.; Mancini, M.; Orlandini, S.; Travaglini, D.; Chirici, G. Exploiting the capabilities of the Sentinel-2 multi spectral instrument for predicting growing stock volume in forest ecosystems. Int. J. Appl. Earth Obs. Geoinf. 2018, 66, 126–134. [Google Scholar] [CrossRef]
- Deo, R.K.; Russell, M.B.; Domke, G.M.; Woodall, C.W.; Falkowski, M.J.; Cohen, W.B. Using Landsat time-series and LiDAR to inform aboveground forest biomass baselines in northern Minnesota, USA. Can. J. Remote Sens. 2017, 43, 28–47. [Google Scholar] [CrossRef]
- Deo, R.; Russell, M.; Domke, G.; Andersen, H.-E.; Cohen, W.; Woodall, C.; Deo, R.K.; Russell, M.B.; Domke, G.M.; Andersen, H.-E.; et al. Evaluating site-specific and generic spatial models of aboveground forest biomass based on Landsat time-series and LiDAR strip samples in the Eastern USA. Remote Sens. 2017, 9, 598. [Google Scholar] [CrossRef]
- McInerney, D.O.; Suarez-Minguez, J.; Valbuena, R.; Nieuwenhuis, M. Forest canopy height retrieval using LiDAR data, medium-resolution satellite imagery and kNN estimation in Aberfoyle, Scotland. Forestry 2010, 83, 195–206. [Google Scholar] [CrossRef] [Green Version]
- Pascual, C.; García-Abril, A.; Cohen, W.B.; Martín-Fernández, S. Relationship between LiDAR-derived forest canopy height and Landsat images. Int. J. Remote Sens. 2010, 31, 1261–1280. [Google Scholar] [CrossRef]
- Maselli, F.; Chiesi, M.; Montaghi, A.; Pranzini, E. Use of ETM+ images to extend stem volume estimates obtained from lidar data. ISPRS J. Photogramm. Remote Sens. 2011, 66, 662–671. [Google Scholar] [CrossRef]
- Wulder, M.A.; White, J.C.; Bater, C.W.; Coops, N.C.; Hopkinson, C.; Chen, G. Lidar plots — a new large-area data collection option: context, concepts, and case study. Can. J. Remote Sens. 2012, 38, 600–618. [Google Scholar] [CrossRef]
- Zald, H.S.J.; Wulder, M.A.; White, J.C.; Hilker, T.; Hermosilla, T.; Hobart, G.W.; Coops, N.C. Integrating Landsat pixel composites and change metrics with lidar plots to predictively map forest structure and aboveground biomass in Saskatchewan, Canada. Remote Sens. Environ. 2016, 176, 188–201. [Google Scholar] [CrossRef] [Green Version]
- Wulder, M.A.; White, J.C.; Nelson, R.F.; Næsset, E.; Ørka, H.O.; Coops, N.C.; Hilker, T.; Bater, C.W.; Gobakken, T. Lidar sampling for large-area forest characterization: A review. Remote Sens. Environ. 2012, 121, 196–209. [Google Scholar] [CrossRef] [Green Version]
- Matasci, G.; Hermosilla, T.; Wulder, M.A.; White, J.C.; Coops, N.C.; Hobart, G.W.; Zald, H.S.J. Large-area mapping of Canadian boreal forest cover, height, biomass and other structural attributes using Landsat composites and lidar plots. Remote Sens. Environ. 2018, 209, 90–106. [Google Scholar] [CrossRef]
- Andersen, H.E.; Strunk, J.; Temesgen, H.; Atwood, D.; Winterberger, K. Using multilevel remote sensing and ground data to estimate forest biomass resources in remote regions: A case study in the boreal forests of interior Alaska. Can. J. Remote Sens. 2012, 37, 596–611. [Google Scholar] [CrossRef]
- Strunk, J.L.; Temesgen, H.; Andersen, H.; Packalen, P. Prediction of Forest Attributes with Field Plots, Landsat, and a Sample of Lidar Strips: A Case Study on the Kenai Peninsula, Alaska. Photogramm. Eng. Remote Sens. 2014, 143–150. [Google Scholar] [CrossRef]
- Saarela, S.; Grafström, A.; Ståhl, G.; Kangas, A.; Holopainen, M.; Tuominen, S.; Nordkvist, K.; Hyyppä, J. Model-assisted estimation of growing stock volume using different combinations of LiDAR and Landsat data as auxiliary information. Remote Sens. Environ. 2015, 158, 431–440. [Google Scholar] [CrossRef]
- Mahoney, C.; Hall, R.; Hopkinson, C.; Filiatrault, M.; Beaudoin, A.; Chen, Q.; Mahoney, C.; Hall, R.J.; Hopkinson, C.; Filiatrault, M.; et al. A forest attribute mapping framework: A pilot study in a Northern boreal forest, Northwest Territories, Canada. Remote Sens. 2018, 10, 1338. [Google Scholar] [CrossRef]
- Boudreau, J.; Nelson, R.F.; Margolis, H.A.; Beaudoin, A.; Guindon, L.; Kimes, D.S. Regional aboveground forest biomass using airborne and spaceborne LiDAR in Québec. Remote Sens. Environ. 2008, 112, 3876–3890. [Google Scholar] [CrossRef]
- Nelson, R.; Boudreau, J.; Gregoire, T.G.; Margolis, H.; Næsset, E.; Gobakken, T.; Ståhl, G. Estimating Quebec provincial forest resources using ICESat/GLAS. Can. J. For. Res. 2009, 39, 862–881. [Google Scholar] [CrossRef]
- Margolis, H.A.; Nelson, R.F.; Montesano, P.M.; Beaudoin, A.; Sun, G.; Andersen, H.-E.; Wulder, M.A. Combining satellite lidar, airborne lidar, and ground plots to estimate the amount and distribution of aboveground biomass in the boreal forest of North America. Can. J. For. Res. 2015, 45, 838–855. [Google Scholar] [CrossRef] [Green Version]
- Nelson, R.; Margolis, H.; Montesano, P.; Sun, G.; Cook, B.; Corp, L.; Andersen, H.-E.; de Jong, B.; Pellat, F.P.; Fickel, T.; et al. Lidar-based estimates of aboveground biomass in the continental US and Mexico using ground, airborne, and satellite observations. Remote Sens. Environ. 2017, 188, 127–140. [Google Scholar] [CrossRef] [Green Version]
- Neigh, C.S.R.; Nelson, R.F.; Ranson, K.J.; Margolis, H.A.; Montesano, P.M.; Sun, G.; Kharuk, V.; Næsset, E.; Wulder, M.A.; Andersen, H.-E. Taking stock of circumboreal forest carbon with ground measurements, airborne and spaceborne LiDAR. Remote Sens. Environ. 2013, 137, 274–287. [Google Scholar] [CrossRef] [Green Version]
- Holm, S.; Nelson, R.; Ståhl, G. Hybrid three-phase estimators for large-area forest inventory using ground plots, airborne lidar, and space lidar. Remote Sens. Environ. 2017, 197, 85–97. [Google Scholar] [CrossRef]
- Hudak, A.T.; Crookston, N.L.; Evans, J.S.; Hall, D.E.; Falkowski, M.J. Nearest neighbor imputation of species-level, plot-scale forest structure attributes from LiDAR data. Remote Sens. Environ. 2008, 112, 2232–2245. [Google Scholar] [CrossRef]
- Hudak, A.T.; Crookston, N.L.; Evans, J.S.; Hall, D.E.; Falkowski, M.J. Corrigendum to “Nearest neighbor imputation of species-level, plot-scale forest structure attributes from LiDAR data”[Remote Sensing of Environment, 112: 2232-2245]. Remote Sens. Environ. 2009, 113, 289–290. [Google Scholar] [CrossRef]
- Penner, M.; Pitt, D.G.; Woods, M.E. Parametric vs. nonparametric LiDAR models for operational forest inventory in boreal Ontario. Can. J. Remote Sens. 2013, 39, 426–443. [Google Scholar] [CrossRef]
- Rowe, J.S. Forest Regions of Canada. Based on W. E. D. Halliday’s “A forest classification for Canada”; Publication No 1300; Department of the Environment, Canadian Forestry Service: Ottawa, ON, Canada, 1972; p. 177. [Google Scholar]
- Government of Newfoundland and Labrador Western Newfoundland Subregions, Forestry and Agrifoods Agency. Available online: https://www.faa.gov.nl.ca/forestry/maps/west_eco.html (accessed on 13 February 2019).
- Canadian Forest Service Canada’s National Forest Inventory Ground Sampling Guidelines. Available online: https://cfs.nrcan.gc.ca/publications?id=29402 (accessed on 23 April 2019).
- Maltamo, M.; Bollandsås, O.M.; Næsset, E.; Gobakken, T.; Packalén, P. Different plot selection strategies for field training data in ALS-assisted forest inventory. Forestry 2011, 84, 23–31. [Google Scholar] [CrossRef]
- Hawbaker, T.J.; Keuler, N.S.; Lesak, A.A.; Gobakken, T.; Contrucci, K.; Radeloff, V.C. Improved estimates of forest vegetation structure and biomass with a LiDAR-optimized sampling design. J. Geophys. Res. Biogeosci. 2009, 114. [Google Scholar] [CrossRef] [Green Version]
- Melville, G.; Stone, C.; Turner, R. Application of LiDAR data to maximise the efficiency of inventory plots in softwood plantations. New Zeal. J. For. Sci. 2015, 45. [Google Scholar] [CrossRef]
- White, J.C.; Wulder, M.A.; Varhola, A.; Vastaranta, M.; Coops, N.C.; Cook, B.D.; Pitt, D.; Woods, M. A Model Development and Application Guide for Generating an Enhanced Forest Inventory Using Airborne Laser Scanning Data and an Area-Based Approach; Information Report FI-X-018; Natural Resources Canada, Canadian Forest Service, Canadian Wood Fibre Centre: Victoria, BC, USA, 2017; p. 50. [Google Scholar]
- Warren, G.R.; Meades, J.P. Wood Defect and Density Studies II Total and Net Volume Equations for Newfoundland Forest Management Units; Information Report N-X-242; Canadian Forestry Service, Newfoundland Forestry Centre: St. John’s, NL, Canada, 1986; p. 14. [Google Scholar]
- Ker, M.F. Metric Yield Tables for the Major Forest Cover Types of Newfoundland; Information Report M-X-141; Natural Resources Canada, Canadian Forest Service, Atlantic Forestry Centre: St. John’s, NL, Canada, 1974; p. 79. [Google Scholar]
- Lambert, M.-C.; Ung, C.-H.; Raulier, F. Canadian national tree aboveground biomass equations. Can. J. For. Res. 2005, 35, 1996–2018. [Google Scholar] [CrossRef]
- ASPRS. Las Specification 1.4 R13; The American Society for Photogrammetry & Remote Sensing: Methesda, Maryland, 2013; p. 28. [Google Scholar]
- Lim Geomatics. LTK LAS Toolkit Version 1.2; Lim Geomatics Inc.: Ottawa, ON, Canada, 2016; p. 32. [Google Scholar]
- Government of Newfoundland and Labrador Forest Types | Forestry and Agrifoods Agency. Available online: https://www.faa.gov.nl.ca/forestry/our_forest/forest_types.html (accessed on 23 April 2019).
- Goetz, S.; Steinberg, D.; Dubayah, R.; Blair, B. Laser remote sensing of canopy habitat heterogeneity as a predictor of bird species richness in an eastern temperate forest, USA. Remote Sens. Environ. 2007, 108, 254–263. [Google Scholar] [CrossRef]
- Van Ewijk, K.Y.; Treitz, P.M.; Scott, N.A. Characterizing forest succession in central Ontario using Lidar-derived indices. Photogramm. Eng. Remote Sens. 2011, 77, 261–269. [Google Scholar] [CrossRef]
- Natural Resources Canada. Canadian Digital Elevation Model: Product Specifications - Edition 1.1; Government of Canada: Sherbrooke, QC, Canada, 2013; p. 11. [Google Scholar]
- Stage, A.R. An expression for the effect of aspect, slope and habitat type on tree growth. For. Sci. 1976, 22, 457–460. [Google Scholar]
- Gatti, A.; Gollapo, A. Sentinel-2 Products Specification Document; Thales Alenia Space: Cannes, France, 2018; p. 487. [Google Scholar]
- Shimada, M.; Itoh, T.; Motooka, T.; Watanabe, M.; Shiraishi, T.; Thapa, R.; Lucas, R. New global forest/non-forest maps from ALOS PALSAR data (2007-2010). Remote Sens. Environ. 2014, 155, 13–31. [Google Scholar] [CrossRef]
- Smith, E.P.; Rose, K.A. Model goodness-of-fit analysis using regression and related techniques. Ecol. Modell. 1995, 77, 49–64. [Google Scholar] [CrossRef]
- Piñeiro, G.; Perelman, S.; Guerschman, J.P.; Paruelo, J.M. How to evaluate models: Observed vs. predicted or predicted vs. observed? Ecol. Modell. 2008, 216, 316–322. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- R Core Team R: A Language and Environment for Statistical Computing. Available online: http://www.r-project.org (accessed on 15 January 2019).
- Freeman, E.A.; Frescino, T.S. Modeling and Map Production using Random Forest and Stochastic Gradient Boosting; USDA Forest Service, Rocky Mountain Research Station: Ogden, UT, USA, 2009; p. 65. [Google Scholar]
- Liaw, A.; Wiener, M. Classification and regression by randomForest. R News 2002, 2, 18–22. [Google Scholar]
- Lumley, T. Based on F. Code by A.M. Leaps: Regression Subset Selection. R Package Version 3.0. Available online: https://cran.r-project.org/package=leaps (accessed on 13 February 2019).
- Mallows, C.L. Some comments on C P. Technometrics 1973, 15, 661–675. [Google Scholar] [CrossRef]
- Myers, R.H. Classical and Modern Regression with Applications, 2nd ed.; PWS-KENT Publishing Company: Boston, MA, USA, 1990; ISBN 0534921787. [Google Scholar]
- Royston, J.P. Algorithm AS 181. Appl. Stat. 2007, 31, 176–180. [Google Scholar] [CrossRef]
- Breusch, T.S.; Pagan, A.R. A simple test for heteroscedasticity and random coefficient variation. Econometrica 1979, 47, 1287–1294. [Google Scholar] [CrossRef]
- Akaike, H. Information Theory and an Extension of the Maximum Likelihood Principle; Petrov, B.N., Csáki, F.C., Eds.; Akademiai Kiado: Budapest, Hungary, 1973. [Google Scholar]
- Hijmans, R.J. Raster: Geographic Data Analysis and Modeling. R Package Version 2.6-7. Available online: https://cran.r-project.org/package=raster (accessed on 13 February 2019).
- ESRI. ArcGIS Desktop: Release 10.4; Environmental Systems Research Institute: Redlands, CA, USA, 2016. [Google Scholar]
- Grafström, A.A.; Lisic, J. BalancedSampling: Balanced and Spatially Balanced Sampling. R Package Version 1.5.2. Available online: https://cran.r-project.org/package=BalancedSampling (accessed on 13 February 2019).
- Grafström, A.; Tillé, Y. Doubly balanced spatial sampling with spreading and restitution of auxiliary totals. Environmetrics 2013, 24, 120–131. [Google Scholar] [CrossRef]
- Grafström, A.; Ringvall, A.H. Improving forest field inventories by using remote sensing data in novel sampling designs. Can. J. For. Res. 2013, 43, 1015–1022. [Google Scholar] [CrossRef]
- Vuong, Q.H. Likelihood ratio tests for model selection and non-nested hypotheses. Econometrica 1989, 57, 307. [Google Scholar] [CrossRef]
- Merkle, E.C.; You, D. Nonnest2: Tests of Non-Nested Models. R Package Version 0.5-2. Available online: https://cran.r-project.org/package=nonnest2 (accessed on 13 February 2019).
- Natural Resources Canada; Public Safety Canada. Federal Airborne LiDAR data Acquisition Guideline Version 1.0. Available online: https://0-doi-org.brum.beds.ac.uk/10.4095/308382 (accessed on 15 February 2019).
- Bolton, D.K.; White, J.C.; Wulder, M.A.; Coops, N.C.; Hermosilla, T.; Yuan, X. Updating stand-level forest inventories using airborne laser scanning and Landsat time series data. Int. J. Appl. Earth Obs. Geoinf. 2018, 66, 174–183. [Google Scholar] [CrossRef]
- Pflugmacher, D.; Cohen, W.B.; Kennedy, R.E.; Yang, Z. Using Landsat-derived disturbance and recovery history and lidar to map forest biomass dynamics. Remote Sens. Environ. 2014, 151, 124–137. [Google Scholar] [CrossRef]
- Vauhkonen, J.; Ørka, H.O.; Holmgren, J.; Dalponte, M.; Heinzel, J.; Koch, B. Tree Species Recognition Based on Airborne Laser Scanning and Complementary Data Sources. In Forestry Applications of Airborne Laser Scanning Concepts and Case Studies; Maltamo, M., Naesset, E., Vauhkonen, J., Eds.; Springer: Dordrecht, The Netherlands, 2014; pp. 135–156. [Google Scholar]
- Tompalski, P.; White, J.C.; Coops, N.C.; Wulder, M.A. Demonstrating the transferability of forest inventory attribute models derived using airborne laser scanning data. Remote Sens. Environ. 2019, 227, 110–124. [Google Scholar] [CrossRef]
- Saarela, S.; Holm, S.; Healey, S.; Andersen, H.-E.; Petersson, H.; Prentius, W.; Patterson, P.; Næsset, E.; Gregoire, T.; Ståhl, G.; et al. Generalized Hierarchical Model-Based Estimation for Aboveground Biomass Assessment Using GEDI and Landsat Data. Remote Sens. 2018, 10, 1832. [Google Scholar] [CrossRef]




| Variable 1 | Units | Calibration (n = 58) | Validation (n = 39) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Min | Max | Mean | Std. Dev. | Min | Max | Mean | Std. Dev. | ||
| HGT | m | 3.4 | 17.1 | 10.1 | 3.6 | 3.7 | 17.0 | 10.5 | 3.6 |
| BA | m2 ha−1 | 0.4 | 63.2 | 30.1 | 18.1 | 0.9 | 62.7 | 33.8 | 16.5 |
| GMV | m3 ha−1 | 0.8 | 401.5 | 156.9 | 118.7 | 1.7 | 376.8 | 175.2 | 109.5 |
| TVOL | m3 ha−1 | 3.6 | 439.9 | 190.1 | 128.6 | 25.1 | 401.0 | 211.7 | 109.5 |
| B | t ha−1 | 6.7 | 260.2 | 121.9 | 67.1 | 33.7 | 214.5 | 132.2 | 52.3 |
| Name | Units | Description | |
|---|---|---|---|
| ALS Variables | |||
| Height Metrics 1 | MAX | m | Maximum height of first returns |
| P95 | m | Height of the 95th percentile of first returns | |
| MEAN | m | Mean height of first returns | |
| Structural Metrics 1 | SKEW | Skewness of first returns | |
| COVAR | Standard deviation of first returns/mean of first returns | ||
| VDR | Vertical distribution ratio [67] | ||
| VCI | Vertical complexity index [68] | ||
| Density Metrics 1 | D2 | % | Percentage of all returns found in bins 1 through 2 of 10 where bin 2 represents the 20th percentile height of all returns |
| D5 | % | Percentage of all returns found in bins 1 through 5 of 10 where bin 5 represents the 50th percentile height of all returns | |
| D8 | % | Percentage of all returns found in bins 1 through 8 of 10 where bin 8 represents the 80th percentile height of all returns | |
| CHM Metrics | CC2 | % | Number of 1 m × 1 m canopy height model cells that have a height value > 2 m divided by the number of nonvoid 1 m × 1 m cells |
| CC6 | % | Number of 1 m × 1 m canopy height model cells that have a height value > 6m divided by the number of nonvoid 1 m × 1 m cells | |
| CC14 | % | Number of 1 m × 1 m canopy height model cells that have a height value > 14m divided by the number of nonvoid 1 m × 1 m cells | |
| Satellite Variables | |||
| Sentinel 2 2 | S2_B2 | % | Blue band; original resolution 10 m |
| S2_B4 | % | Red band; original resolution 10 m | |
| S2_B5 | % | Vegetation red edge; resolution 20 m | |
| S2_B8 | % | NIR; original resolution 10 m | |
| S2_B11 | % | SWIR; resolution 20 m | |
| PALSAR | HH | DN | Radar backscatter — HH polarization; original resolution 25 m |
| HV | DN | Radar backscatter — HV polarization; original resolution 25 m | |
| Environmental Variables | |||
| Topographic and Solar Radiation | Elevation | m | Elevation above mean sea level from 0.75 arc second CDEM [69] |
| CosAspect | −1 to 1 | cos(Aspect) transformation representing northness | |
| SCOSA | Slope × cos(Aspect) transformation [70] | ||
| SINA | Slope × sin(Aspect) transformation [70] | ||
| Random Forest | Regression | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| R2 | RMSD | RMSD% | Bias | Bias% | R2 | RMSD | RMSD% | Bias | Bias% | |
| Calibration 2 (n = 58) | ||||||||||
| HGT | 0.93 | 0.95 | 9.37 | 0.09 | 0.84 | 0.95 | 0.84 | 8.27 | 0.00 | 0.00 |
| BA | 0.90 | 5.67 | 18.86 | 0.08 | 0.28 | 0.92 | 5.22 | 17.37 | 0.00 | 0.00 |
| GMV | 0.94 | 29.11 | 18.56 | 1.35 | 0.86 | 0.94 | 30.31 | 19.33 | 0.00 | 0.00 |
| TVOL | 0.94 | 31.29 | 16.46 | 0.04 | 0.02 | 0.96 | 27.72 | 14.58 | 0.00 | 0.00 |
| B | 0.90 | 21.49 | 17.63 | 0.43 | 0.35 | 0.93 | 18.74 | 15.37 | 0.00 | 0.00 |
| Validation (n = 39) | ||||||||||
| HGT | 0.94 | 1.14 | 10.86 | −0.53 | −5.09 | 0.95 | 1.03 | 9.79 | −0.54 | −5.17 |
| BA | 0.83 | 7.56 | 22.40 | −3.03 | −8.97 | 0.86 | 6.71 | 19.87 | −2.44 | −7.23 |
| GMV | 0.88 | 44.76 | 25.55 | −22.19 | −12.67 | 0.90 | 40.07 | 22.87 | −18.40 | −10.50 |
| TVOL | 0.91 | 37.84 | 17.87 | −17.03 | −8.04 | 0.91 | 37.21 | 17.57 | −13.32 | −6.29 |
| B | 0.85 | 22.14 | 16.75 | −5.86 | −4.43 | 0.83 | 23.71 | 17.93 | −4.28 | −3.24 |
| Random Forest | Regression | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| R2 | RMSD | RMSD% | Bias | Bias% | R2 | RMSD | RMSD% | Bias | Bias% | |
| Calibration 2 (n = 5000) | ||||||||||
| HGT | 0.71 | 1.81 | 17.85 | 0.01 | 0.09 | 0.68 | 2.17 | 21.16 | 0.00 | 0.00 |
| BA | 0.70 | 8.14 | 27.11 | 0.04 | 0.14 | 0.66 | 9.80 | 32.20 | 0.00 | 0.00 |
| GMV | 0.67 | 59.40 | 31.66 | 0.50 | 0.26 | 0.58 | 73.06 | 36.49 | 0.00 | 0.00 |
| TVOL | 0.70 | 61.64 | 28.30 | 0.14 | 0.06 | 0.61 | 76.73 | 34.73 | 0.00 | 0.00 |
| B | 0.74 | 30.43 | 23.31 | 0.21 | 0.16 | 0.67 | 39.43 | 30.63 | 0.00 | 0.00 |
| Validation (n = 39) | ||||||||||
| HGT | 0.84 | 1.66 | 15.88 | −0.09 | −0.91 | 0.79 | 1.74 | 16.59 | 0.31 | 2.94 |
| BA | 0.76 | 8.87 | 26.27 | −2.39 | −7.08 | 0.64 | 9.93 | 29.43 | −0.75 | −2.24 |
| GMV | 0.79 | 55.14 | 31.47 | −3.04 | −1.74 | 0.67 | 63.41 | 36.20 | 10.13 | 5.78 |
| TVOL | 0.74 | 56.55 | 26.71 | −6.60 | −3.11 | 0.64 | 66.66 | 31.48 | 7.64 | 3.61 |
| B | 0.65 | 31.36 | 23.72 | −3.07 | −2.32 | 0.56 | 37.15 | 28.10 | 4.47 | 3.38 |
| (a) Direct vs. Indirect | ||||||||||
| Random Forest | Regression | |||||||||
| R2DIR | R2IND | pDIFF | pDIRvsIND | pINDvsDIR | R2DIR | R2IND | pDIFF | pDIRvsIND | pIND vsDIR | |
| HGT | 0.67 | 0.86 | 0.000 | 0.999 | 0.001 | 0.70 | 0.79 | 0.001 | 0.996 | 0.004 |
| BA | 0.68 | 0.77 | 0.000 | 0.947 | 0.053 | 0.54 | 0.65 | 0.000 | 0.982 | 0.018 |
| GMV | 0.67 | 0.80 | 0.000 | 0.977 | 0.023 | 0.50 | 0.68 | 0.000 | 1.000 | 0.000 |
| TVOL | 0.54 | 0.76 | 0.000 | 0.993 | 0.007 | 0.54 | 0.65 | 0.000 | 0.960 | 0.040 |
| B | 0.44 | 0.66 | 0.000 | 0.991 | 0.009 | 0.47 | 0.56 | 0.000 | 0.895 | 0.105 |
| (b) Random Forest vs. Regression | ||||||||||
| Direct | Indirect | |||||||||
| R2RF | R2REG | pDIFF | pRFvsREG | pREGvsRF | R2RF | R2REG | pDIFF | pRFvsREG | pREGvsRF | |
| HGT | 0.67 | 0.70 | 0.000 | 0.626 | 0.374 | 0.86 | 0.79 | 0.000 | 0.025 | 0.975 |
| BA | 0.68 | 0.54 | 0.000 | 0.060 | 0.940 | 0.77 | 0.65 | 0.000 | 0.000 | 1.000 |
| GMV | 0.67 | 0.50 | 0.000 | 0.018 | 0.982 | 0.80 | 0.68 | 0.000 | 0.004 | 0.996 |
| TVOL | 0.54 | 0.54 | 0.000 | 0.491 | 0.509 | 0.76 | 0.65 | 0.000 | 0.002 | 0.998 |
| B | 0.44 | 0.47 | 0.000 | 0.617 | 0.383 | 0.66 | 0.56 | 0.000 | 0.008 | 0.992 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luther, J.E.; Fournier, R.A.; van Lier, O.R.; Bujold, M. Extending ALS-Based Mapping of Forest Attributes with Medium Resolution Satellite and Environmental Data. Remote Sens. 2019, 11, 1092. https://0-doi-org.brum.beds.ac.uk/10.3390/rs11091092
Luther JE, Fournier RA, van Lier OR, Bujold M. Extending ALS-Based Mapping of Forest Attributes with Medium Resolution Satellite and Environmental Data. Remote Sensing. 2019; 11(9):1092. https://0-doi-org.brum.beds.ac.uk/10.3390/rs11091092
Chicago/Turabian StyleLuther, Joan E., Richard A. Fournier, Olivier R. van Lier, and Mélodie Bujold. 2019. "Extending ALS-Based Mapping of Forest Attributes with Medium Resolution Satellite and Environmental Data" Remote Sensing 11, no. 9: 1092. https://0-doi-org.brum.beds.ac.uk/10.3390/rs11091092