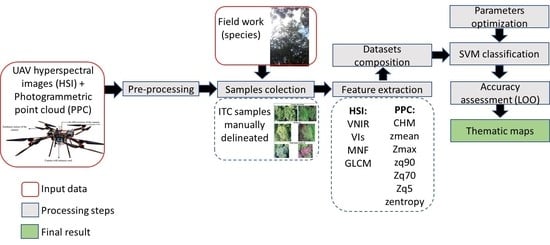
5.1. Effects of Feature Selection and Data Fusion
This study showed that the combination of UAV-PPC features with the VNIR bands led to a significant increase in tree species classification accuracies. The best result was achieved with the VNIR_PPC_CHM_VIs dataset, with 72.4% of OA, significantly better when compared with the use of VNIR bands alone (57% of OA). The first work involving the investigation of UAV-based photogrammetry and hyperspectral imaging in individual tree detection and tree species classification was made by Nevalainen et al. [
23], in which they tested features extracted from UAV hyperspectral data and from PPC to classify tree species in a boreal forest, achieving 95% of OA. Contrary to our study, they did not find any improvement with the addition of PPC features. On the other hand, Tuominen et al. [
24] tested the use of UAV hyperspectral data and PPC features to classify 26 tree species in an arboretum located in Finland and reported an increase of 0.07 in Kappa index when they combined the VNIR bands with 3D features (Kappa of 0.77). The aforementioned studies reached better or similar accuracies to our study, but none of them involved tropical environments. Nevalainen et al. [
23] only classified four tree species in a boreal forest and, despite the high number of tree species, the study of Tuominen et al. [
24] was conducted in an arboretum area with the tree species distribution in homogeneous stands, which makes the area suitable for testing tree species recognition both at the stand and individual tree level. Our study is the first study involving this type of data to classify a high number of tree species in a subtropical forest.
Despite the study made by Nevalainen et al. [
23] did not find improvements with the addition of PPC features, the importance of using 3D information, such as that derived from LiDAR, for tree species classification have been reported in several other studies. Deng et al. [
89] performed the classification of tree species in a temperate forest and obtained an improvement of up 14% when the LiDAR-derived features were employed together with an RGB ortophoto. Shen and Cao [
19] had an improvement of 0.4% to 5.6% when using both hyperspectral and airborne LiDAR features to classify tree species in a subtropical forest. Similarly, Piiroinen et al. [
55] found an increase of 6% when LiDAR features were added to the hyperspectral bands for classifying tree species in a diverse African agroforestry. Dalponte et al. [
11] and Jones et al. [
35] reported that LiDAR combined with hyperspectral data increased the classification accuracy of ~2%, with a greater improvement for some tree species. It can also be noticed in this study that the improvement was more evident for some species, such as
Ocotea sp./
Nectandra megapotamica and
Luehea divaricata/
Cedrela fissilis.
Ocotea sp. class comprises two species of
Ocotea genus (
O. pulchella and
O. puberula), which result in a more diverse spectral behavior. Furthermore
Ocotea sp. and
Nectandra megapotamica belong to the same family (Lauraceae), having similar spectral characteristics. In this case, the spectral similarity among these tree species can be solved with the inclusion of PPC features.
Luehea divaricata and
Cedrela fissilis can reach similar heights, thus, the addition of CHM band was not as useful as the inclusion of PPC features to discriminate them. In this case, the PPC features may captured differences in their crown structure, since the former has a wider and denser top with irregular branching, while the latter has a smaller and rounded top, with forked branching [
87].
The inclusion of the height information (CHM) also improved accuracies, but to a lesser extent than PPC features: approximately 4% compared with the use of the VNIR bands alone. In the studies of Cho et al. [
90], Naidoo et al. [
91] and Asner et al. [
92], the tree height derived from LiDAR was an important variable for mapping tree species in two completely different forest ecosystems (savannas and tropical forest). On the other hand, Ghosh et al. [
12] concluded that there is no significant effect of the height information on tree species classification accuracies in a temperate forest. According to Fassnacht et al. [
13], the canopy height per se is not a good predictor to classify tree species as the height of a tree varies with age, site conditions, and competition, and only to a minor degree with species. However, in tropical forests, this predictor can be useful to discriminate species belonging to different successional groups as pioneer, secondary, and climactic, since they tend to have different heights. In this study, there was a great confusion between
Mimosa scabrella and
Podocarpus lambertii in the datasets without the CHM information.
Mimosa scabrella is considered a pioneer species, reaching between 4 and 18 m.
Podocarpus lambertii, a late secondary tree, usually is taller, reaching more than 20 m [
89]. For this reason, when the height information was incorporated, there was an increase in accuracies of these species, mainly for
Mimosa scabrella.
Araucaria angustifolia, the most frequent tree in the study area, presented stable accuracies even with the inclusion of 3D information, showing that even the use of the VNIR bands alone can discriminate this tree species. It can be observed that this species presents the lowest radiance values in all spectra of FPI bands (
Figure 3). According to Roberts et al. [
93], coniferous trees generally have lower reflectance values in the NIR region compared to broadleaves trees, which is closely related to their needle structure and the higher absorption of coniferous needles. Furthermore, crown size and shape of coniferous trees influence the hemispherical directional reflectance factor (HDRF) and thus their reflectance as well [
94]. On the other hand,
Podocarpus lambertii, another conifer of the area, tended to be more confused with some broadleaf species. This may have occurred because its shape is not conic as a normal pine, and its branch structure led to confusion with broadleaves species with small leaves, such as
Mimosa scabrella.
All the VNIR bands showed similar importance according to JM distance (
Figure 4). Only slightly higher importance values can be noticed for four bands between 659 and 690 nm. This region included the chlorophyll absorption features, previously reported to contain useful information for the separation of tree species with hyperspectral data [
18,
54]. The NIR bands, pointed as an important region for tree species classification [
15,
37], did not stand out in this study. In this region, the tree structure has the strongest impact [
95] and the changes in view angle may reduce the relative spectral differences between the species [
23]. Furthermore, due to the limited spectral range of the FPI camera (506–819 nm), the complete infrared region (including SWIR) could not be fully tested and assessed.
The inclusion of hyperspectral features (MNF, GLCM, and VIs) in the VNIR_PPC_CHM dataset did not bring significant improvement in accuracy, or even worsened with GLCM features. When all of them were added to the dataset composed solely by VNIR bands (VNIR_VIs_MNF_GLCM) an increase of 1.7% in OA was observed. This increase was also observed when VIs were added to the MNF_PPC_CHM dataset. Ferreira et al. [
18] reported an increase in the classification accuracy (of up to 5%) adding the VIs to the VNIR dataset. Maschler et al. [
22] found great improvement in tree species classification accuracies when VIs and principal components were aggregated to the classifications, while the inclusion of textural metrics only had a small effect. Although it was not the most important spectral region according to the JM distance, the VIs containing at least one NIR band (PSSR and NDVI) were more important than PSRI and PRI for tree species classification. In the study of Naidoo et al. [
93], the NDVI was also scored as one of the most important VIs to classify eight savanna tree species with hyperspectral data. Some studies suggested that band ratios and VIs generated using different band combinations [
18,
38,
89,
91] have advantages for species differentiation and biomass estimation because these features can reduce bidirectional reflectance distribution function (BRDF) errors and do not saturate as quickly as single band data [
96]. The use of MNF features (MNF_CHM_PPC dataset) instead of VNIR bands (VNIR_CHM_PPC dataset) led to a decrease of 3.1% in OA. Ghosh et al. [
12] and Piiroinen et al. [
55] reported an improvement when MNF components were used instead of spectral bands, but it is worth noting that in those cases they had hyperspectral data with more than 100 bands. In those cases, MNF transformation reduced the dimensionality and redundancy inherent in high spectral resolution data and, thus, provided better accuracies [
12]. Nevertheless, in this study the first four MNF features showed equal or even higher JM values than the VNIR bands.
The use of all VNIR bands, hyperspectral features and CHM and PPC features in the
full dataset did not improve the accuracy when compared with the datasets involving the VNIR, PPC, and CHM. In the FS process 48 of the 68 features were selected, resulting in a nonsignificant increase in accuracy when compared with the
full dataset. It should be pointed out that studies that performed the FS in hyperspectral data for tree species classification, generally dealt with more than 100 spectral bands [
18,
22,
37,
54,
55]. However, our hyperspectral data contain only 25 bands, and thus the spectral information is not as redundant as when more than 100 hyperspectral bands are used. Furthermore, according to Fassnacht et al. [
13], the FS is commonly applied to reduce the processing time and to enable a meaningful interpretation of the selected predictors, not necessarily resulting in significant increases in classification accuracies. Deng et al. [
89], when testing a FS process for tree species classification, stated that for the same feature there were different contribution degrees to species classification in different loops, indicating that the importance of a feature is changeable and greatly depends on the combination with other features. Therefore, it is difficult to determine a combination of features that can benefit all tree species classes at the same time.
5.2. Consideration about the Number of Samples, Tree Species Classes, and Classification Method
Most previous studies concerning tree species classification using UAV (as well as airborne) datasets have been carried out in forests with less-diverse species structure, like boreal and temperate forests (e.g., [12,22,23,37,38,40,56,61,89]), in which is common to find accuracies over 85% (e.g., [
12,
38,
40,
89]). However, our findings are consistent with other studies involving airborne hyperspectral data for tree species classification in tropical forest environments. Féret and Asner [
21] reached an OA of 73.2% using the pixel-based approach to classify 17 tree species in a Hawaiian tropical forest with airborne hyperspectral data, and 74.9% when they classified only 10 species. When testing machine learning algorithms to classify eight tree species of a subtropical forest in Brazil, Ferreira et al. [
18] achieved an average classification accuracy of 70% using the VNIR hyperspectral bands. In that case, the inclusion of shortwave infrared (SWIR) bands increased the accuracy to 84%. Using hyperspectral data with visible, NIR and SWIR bands, Clark and Roberts [
15] reached an OA of 71.5% in the pixel-based approach to classify seven emerged-canopy species in a tropical forest in Costa Rica. Regarding studies involving other high diverse environments, Piironen et al. [
55] reached 57% of OA when classifying 31 tree species in a diverse agroforestry landscape in Africa. Graves et al. [
97] classified 20 tree species and one mixed-species class in a tropical agricultural landscape in Panama and reported an OA of 62%.
According to Ferét and Asner [
21], the minimum number of samples per species required to perform optimal classification is a limiting factor in tropical forest studies due to the difficulty and high cost of tree localization on the ground. Our study, similarly to other studies involving the classification of a considerable number of tree species, e.g., Féret and Asner [
21], Tuominen et al. [
24] and Piiroinen et al. [
55], had only few ITC samples for some classes. This, coupled with the fact that not all the species were considered in the SVM model, is an obstacle to extrapolate the classification model over the entire area, since it may result in a map with many uncertainties (
Figure 6). Even so, this kind of map could be used in some general ecological applications such as assessing patterns of species composition and abundance across environmental gradients or land management units, identification of areas of high or low tree cover and species diversity, identification of ecological groups of tree species (as pioneer, secondary, and climax) assisting the successional forest stage classification, and providing landscape estimates of aboveground biomass [
97]. For more focused applications where accurate predictions of species location and identity is needed, such as monitoring endangered tree species (e.g.,
Araucaria angustifolia and
Cedrela fissilis), classification and mapping errors may be too large. For these applications, techniques such as semisupervised methods where a focal group of species is identified from a background of unknown species [
16,
98], can be a better approach [
97].
The SVM classifier proved to be a suitable choice for this study, maintaining a relatively stable performance with few samples, even when the complexity of the datasets increased with the addition of several features. This algorithm has proved to be a promising approach for tree species classification, having a similar [
12,
40] or even surpassing the Random Forest algorithm performance [
18,
37,
55,
56,
89,
99]. Some studies showed that SVM performs better than RF in the presence of small or imbalanced datasets. In this situation, RF tends to focus more on the prediction accuracy of the majority class, which often results in poor accuracy for the minority classes [
37,
100]. On the other hand, RF has the advantage of having fewer parameters to be tuned, a lower computational cost [
37] and it is less affected by correlated variables [
15].
This work adopted a pixel-based approach for tree species level classification. According to Heinzel and Koch [
36], object-based approaches using individual ITCs as classification units may reduce the negative effects of spectral variability of pixels. However, most of studies that indicated advantages in object-based approaches did not use the ITC as a classification unit, but instead grouped the pixels into segments after the classification, using a majority class rule procedure [
18,
20,
21]. Clark and Roberts [
15] compared the object-based approach, composed by the mean spectra of each ITC, with pixel and majority rule approaches. The pixel classification had roughly the same performance (~70% OA) as when using the average spectra from ITCs, however the pixel-majority classification had much better accuracy, 87%. Féret and Asner [
21] reached better results in an object-based approach (79.6%) when compared with the pixel classification (74.9% of OA) to classify tree species in a tropical forest. However, for the object-based classification, they had to reduce the number of classes from 17 to 10, due to the limited number of pixels and the tree crowns labeled to train the classifiers. Furthermore, according to the authors, the segmentation did not delineate ITCs correctly, but each crown was composed by several segments.
Indeed, a proper ITC delineation in a high diversity tropical forest is a complicated task, because of its complex structure. In this sense, most of the published studies focusing on ITC delineation are concentrated in coniferous forests, since most algorithms assume a basic conical crown shape, which is more appropriate for conifers [
101]. If an object-based methodology can integrate spectral information with crown-shape parameters, it will greatly assist the process of tree mapping and more accurately fit the needs of tropical forest ecosystem management [
10,
67]. Therefore, the production of reliable species maps in a high diversity tropical forest with object-based approaches is still a topic to be explored.