Remotely Sensed Variables of Ecosystem Functioning Support Robust Predictions of Abundance Patterns for Rare Species
Abstract
:1. Introduction
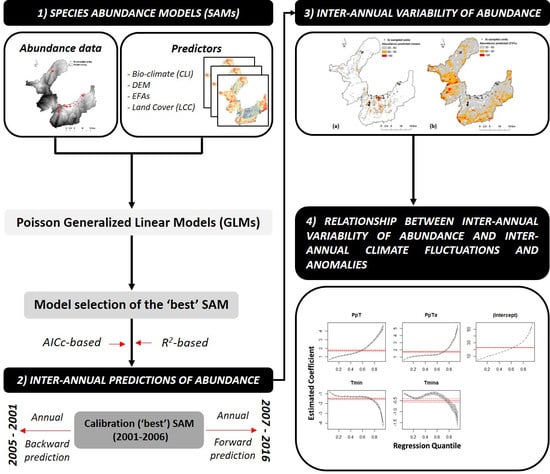
2. Materials and Methods
2.1. Test Species and Study Area
2.2. Species Abundance Dataset
2.3. Modeling Framework
2.3.1. Environmental Predictors
2.3.2. Model Fitting
2.4. Inter-Annual Predictions of Iris Abundance
3. Results
3.1. Ranking of Models and Predictors
3.2. Inter-Annual Variability of Iris Abundance
4. Discussion
4.1. Ecosystem Functioning Attributes as Predictors in Species Abundance Models
4.2. Inter-Annual Variability of Species Abundance and its Driving Forces
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- IPBES. The Intergovernmental Science-Policy Platform on Biodiversity and Ecosystem Services; IPBES: Paris, France, 2019. [Google Scholar]
- Pereira, H.M.; Ferrier, S.; Walters, M.; Geller, G.N.; Jongman, R.H.G.; Scholes, R.J.; Bruford, M.W.; Brummitt, N.; Butchart, S.H.M.; Cardoso, A.C.; et al. Essential Biodiversity Variables. Science 2013, 339, 277–278. [Google Scholar] [CrossRef] [Green Version]
- Pereira, H.; Belnap, J.; Böhm, M.; Brummitt, N.; Garcia-Moreno, J.; Gregory, R.; Martin, L.; Peng, C.; Swaay, V.P.; Schmeller, D.; et al. Monitoring essential biodiversity variables at the species level. In The GEO Handbook on Biodiversity Observation Networks; Walters, M., Scholes, R., Eds.; Springer: Berlin/Heidelberg, Germany, 2017. [Google Scholar]
- Kissling, W.D.; Ahumada, J.A.; Bowser, A.; Fernandez, M.; Fernández, N.; García, E.A.; Guralnick, R.P.; Isaac, N.J.B.; Kelling, S.; Los, W.; et al. Building essential biodiversity variables (EBVs) of species distribution and abundance at a global scale. Biol. Rev. 2018, 93, 600–625. [Google Scholar] [CrossRef]
- Nielsen, S.E.; Johnson, C.J.; Heard, D.C.; Boyce, M.S. Can models of presence-absence be used to scale abundance? Two case studies considering extremes in life history. Ecography 2005, 28, 197–208. [Google Scholar] [CrossRef]
- Elith, J.; Leathwick, J.R. Species distribution models: ecological explanation and prediction across space and time. Annu. Rev. Ecol. Evol. Syst. 2009, 40, 677–697. [Google Scholar] [CrossRef]
- Van Couwenberghe, R.; Collet, C.; Pierrat, J.-C.; Verheyen, K.; Gégout, J.-C. Can species distribution models be used to describe plant abundance patterns? Ecography 2013, 36, 665–674. [Google Scholar] [CrossRef]
- Scholes, R.J.; Walters, M.; Turak, E.; Saarenmaa, H.; Heip, C.H.; Tuama, É.Ó.; Faith, D.P.; Mooney, H.A.; Ferrier, S.; Jongman, R.H.; et al. Building a global observing system for biodiversity. Curr. Opin. Environ. Sustain. 2012, 4, 139–146. [Google Scholar] [CrossRef]
- Tittensor, D.P.; Walpole, M.; Hill, S.L.L.; Boyce, D.G.; Britten, G.L.; Burgess, N.D.; Butchart, S.H.M.; Leadley, P.W.; Regan, E.C.; Alkemade, R.; et al. A mid-term analysis of progress toward international biodiversity targets. Science 2014, 346, 241–244. [Google Scholar] [CrossRef]
- Honrado, J.P.; Pereira, H.M.; Guisan, A. Fostering integration between biodiversity monitoring and modelling. J. Appl. Ecol. 2016, 53, 1299–1304. [Google Scholar] [CrossRef]
- Araújo, M.B.; Guisan, A. Five (or so) challenges for species distribution modelling. J. Biogeogr. 2006, 33, 1677–1688. [Google Scholar] [CrossRef]
- Zimmermann, N.E.; Edwards, T.C., Jr.; Graham, C.H.; Pearman, P.B.; Svenning, J.-C. New trends in species distribution modelling. Ecography 2010, 33, 985–989. [Google Scholar] [CrossRef]
- Howard, C.; Stephens, P.A.; Pearce-Higgins, J.W.; Gregory, R.D.; Willis, S.G. Improving species distribution models: the value of data on abundance. Methods Ecol. Evol. 2014, 5, 506–513. [Google Scholar] [CrossRef] [Green Version]
- Ehrlén, J.; Morris, W.F. Predicting changes in the distribution and abundance of species under environmental change. Ecol. Lett. 2015, 18, 303–314. [Google Scholar] [CrossRef]
- Jetz, W.; McGeoch, M.A.; Guralnick, R.; Ferrier, S.; Beck, J.; Costello, M.J.; Fernandez, M.; Geller, G.N.; Keil, P.; Merow, C.; et al. Essential biodiversity variables for mapping and monitoring species populations. Nat. Ecol. Evol. 2019, 3, 539–551. [Google Scholar] [CrossRef] [Green Version]
- Ives, A.R. Predicting the response of populations to environmental change. Ecology 1995, 76, 926–941. [Google Scholar] [CrossRef]
- Mutshinda, C.M.; O’Hara, R.B.; Woiwod, I.P. What drives community dynamics? Proc. R. Soc. B Biol. Sci. 2009, 276, 2923–2929. [Google Scholar] [CrossRef] [Green Version]
- Mutshinda, C.M.; O’Hara, R.B.; Woiwod, I.P. A Multispecies perspective on ecological impacts of climatic forcing. J. Anim. Ecol. 2011, 80, 101–107. [Google Scholar] [CrossRef]
- Oliver, T.H.; Morecroft, M.D. Interactions between climate change and land use change on biodiversity: Attribution problems, risks, and opportunities. Wiley Interdiscip. Rev. Clim. Chang. 2014, 5, 317–335. [Google Scholar] [CrossRef]
- Lomolino, M.V.; Riddle, B.R.; Whittaker, R.J. (Eds.) Biogeography, 5th ed.; Oxford University Press: Sunderland, MA, USA, 2017. [Google Scholar]
- He, K.S.; Bradley, B.A.; Cord, A.F.; Rocchini, D.; Tuanmu, M.-N.; Schmidtlein, S.; Turner, W.; Wegmann, M.; Pettorelli, N. Will remote sensing shape the next generation of species distribution models? Remote Sens. Ecol. Conserv. 2015, 1, 4–18. [Google Scholar] [CrossRef] [Green Version]
- Pettorelli, N.; to Bühne, H.; Tulloch, A.; Dubois, G.; Macinnis-Ng, C.; Queirós, A.M.; Keith, D.A.; Wegmann, M.; Schrodt, F.; Stellmes, M.; et al. Satellite remote sensing of ecosystem functions: Opportunities, challenges and way forward. Remote Sens. Ecol. Conserv. 2018, 4, 71–93. [Google Scholar] [CrossRef]
- Jax, K. (Ed.) Ecosystem Functioning; Cambridge University Press: Cambridge, UK, 2010. [Google Scholar] [CrossRef]
- Cabello, J.; Fernández, N.; Alcaraz-Segura, D.; Oyonarte, C.; Piñeiro, G.; Altesor, A.; Delibes, M.; Paruelo, J.M. The ecosystem functioning dimension in conservation: Insights from remote sensing. Biodivers. Conserv. 2012, 21, 3287–3305. [Google Scholar] [CrossRef]
- Mouillot, D.; Graham, N.A.J.; Villéger, S.; Mason, N.W.H.; Bellwood, D.R. A functional approach reveals community responses to disturbances. Trends Ecol. Evol. 2013, 28, 167–177. [Google Scholar] [CrossRef]
- Pettorelli, N.; Wegmann, M.; Skidmore, A.; Mücher, S.; Dawson, T.P.; Fernandez, M.; Lucas, R.; Schaepman, M.E.; Wang, T.; O’Connor, B.; et al. Framing the concept of satellite remote sensing essential biodiversity variables: Challenges and future directions. Remote Sens. Ecol. Conserv. 2016, 2, 122–131. [Google Scholar] [CrossRef]
- Alcaraz-Segura, D.; Lomba, A.; Sousa-Silva, R.; Nieto-Lugilde, D.; Alves, P.; Georges, D.; Vicente, J.R.; Honrado, J.P. Potential of satellite-derived ecosystem functional attributes to anticipate species range shifts. Int. J. Appl. Earth Obs. Geoinf. 2017, 57, 86–92. [Google Scholar] [CrossRef]
- Regos, A.; Gagne, L.; Alcaraz-Segura, D.; Honrado, J.P.; Domínguez, J. Effects of species traits and environmental predictors on performance and transferability of ecological niche models. Sci. Rep. 2019, 9, 4221. [Google Scholar] [CrossRef]
- Requena-Mullor, J.M.; López, E.; Castro, A.J.; Alcaraz-Segura, D.; Castro, H.; Reyes, A.; Cabello, J. Remote-sensing based approach to forecast habitat quality under climate change scenarios. PLoS ONE 2017, 12, e0172107. [Google Scholar] [CrossRef]
- Arenas-Castro, S.; Goncalves, J.; Alves, P.; Alcaraz-Segura, D.; Honrado, J.P. Assessing the multi-scale predictive ability of ecosystem functional attributes for species distribution modelling. PLoS ONE 2018. [Google Scholar] [CrossRef]
- Ortiz, S.; Pulgar Sañudo, I. Iris Boissieri. In The IUCN Red List of Threatened Species 2011; IUCN: Cambridge, UK, 2011. [Google Scholar]
- Evans, D.; Arvela, M. Assessment and reporting under article 17 of the habitats directive. In Explanatory Notes & Guidelines for the period 2007–2012; European Commission: Brussels, Belgium, 2011. [Google Scholar]
- Plano de Ordenamento Do Parque Nacional Da Peneda-Gerês. 2010. Available online: http://www2.icnf.pt/portal/florestas/dfci/inc/info-geo (accessed on 5 August 2019).
- Ninyerola, M.; Pons, X.; Roure, J.M. A methodological approach of climatological modelling of air temperature and precipitation through GIS techniques. Int. J. Climatol. 2000, 20, 1823–1841. [Google Scholar] [CrossRef]
- Gorelick, N.; Hancher, M.; Dixon, M.; Ilyushchenko, S.; Thau, D.; Moore, R. Google earth engine: Planetary-scale geospatial analysis for everyone. Remote Sens. Environ. 2017, 202, 18–27. [Google Scholar] [CrossRef]
- Alcaraz-Segura, D.; Cabello, J.; Paruelo, J.M.; Delibes, M. Use of descriptors of ecosystem functioning for monitoring a national park network: A remote sensing approach. Environ. Manag. 2009, 43, 38–48. [Google Scholar] [CrossRef]
- Guisan, A.; Edwards, T.C.; Hastie, T. Generalized linear and generalized additive models in studies of species distributions: Setting the scene. Ecol. Modell. 2002, 157, 89–100. [Google Scholar] [CrossRef]
- O’Hara, R.B.; Kotze, D.J. Do not log-transform count data. Methods Ecol. Evol. 2010, 1, 118–122. [Google Scholar] [CrossRef] [Green Version]
- St-Pierre, A.P.; Shikon, V.; Schneider, D.C. Count data in biology-data transformation or model reformation? Ecol. Evol. 2018, 8, 3077–3085. [Google Scholar] [CrossRef]
- Symonds, M.R.; Moussalli, A. A brief guide to model selection, multimodel inference and model averaging in behavioural ecology using akaike’s information criterion. Behav. Ecol. Sociobiol. 2011, 65, 13–21. [Google Scholar] [CrossRef]
- Burnham, K.; Anderson, D. Model Selection and Multimodel Inference: A Practical Information-Theoretic Approach; Springer: Berlin/Heidelberg, Germany, 2002. [Google Scholar]
- Hyndman, R.J.; Koehler, A.B. Another look at measures of forecast accuracy. Int. J. Forecast. 2006, 22, 679–688. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.-Y.; Syu, H.-S. A comparative study with quantile regression and back propagation neural network for credit rating. J. Financ. Econ. 2016, 4, 46–57. [Google Scholar] [CrossRef]
- Informação Georeferenciada das áreas Ardidas. Available online: http://www2.icnf.pt/portal/florestas/dfci/inc/info-geo (accessed on 5 August 2019).
- Abatzoglou, J.T.; Dobrowski, S.Z.; Parks, S.A.; Hegewisch, K.C. TerraClimate, a High-Resolution Global Dataset of Monthly Climate and Climatic Water Balance from 1958–2015. Sci. Data 2018, 5, 170191. [Google Scholar] [CrossRef]
- Wolda, H.; Marek, J. Measuring variation in abundance, the problem with zeros. Eur. J. Entomol. 1994, 91, 145–161. [Google Scholar]
- Fletcher, D.; MacKenzie, D.; Villouta, E. Modelling skewed data with many zeros: A simple approach combining ordinary and logistic regression. Environ. Ecol. Stat. 2005, 12, 45–54. [Google Scholar] [CrossRef]
- Koenker, R.; Machado, J.A.F. Goodness of fit and related inference processes for quantile regression. J. Am. Stat. Assoc. 1999, 94, 1296–1310. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019. [Google Scholar]
- Gonçalves, J.; Alves, P.; Pôças, I.; Marcos, B.; Sousa-Silva, R.; Lomba, Â.; Honrado, J.P. Exploring the spatiotemporal dynamics of habitat suitability to improve conservation management of a vulnerable plant species. Biodivers. Conserv. 2016, 25, 2867–2888. [Google Scholar] [CrossRef]
- Sirami, C.; Caplat, P.; Popy, S.; Clamens, A.; Arlettaz, R.; Jiguet, F.; Brotons, L.; Martin, J.-L. Impacts of global change on species distributions: obstacles and solutions to integrate climate and land use. Glob. Ecol. Biogeogr. 2017, 26, 385–394. [Google Scholar] [CrossRef]
- Raven, P.H. (Ed.) Predicting Species Occurrences: Issues of Accuracy and Scale; Island Press: Washington, DC, USA, 2002. [Google Scholar]
- Dallas, T.A.; Hastings, A. Habitat suitability estimated by niche models is largely unrelated to species abundance. Glob. Ecol. Biogeogr. 2018, 27, 1448–1456. [Google Scholar] [CrossRef]
- Pearson, R.G.; Dawson, T.P. Predicting the impacts of climate change on the distribution of species: are bioclimate envelope models useful? Glob. Ecol. Biogeogr. 2003, 12, 361–371. [Google Scholar] [CrossRef] [Green Version]
- Deblauwe, V.; Droissart, V.; Bose, R.; Sonké, B.; Blach-Overgaard, A.; Svenning, J.-C.; Wieringa, J.J.; Ramesh, B.R.; Stévart, T.; Couvreur, T.L.P. Remotely sensed temperature and precipitation data improve species distribution modelling in the tropics. Glob. Ecol. Biogeogr. 2016, 25, 443–454. [Google Scholar] [CrossRef]
- Lassueur, T.; Joost, S.; Randin, C.F. Very high resolution digital elevation models: Do they improve models of plant species distribution? Ecol. Modell. 2006, 198, 139–153. [Google Scholar] [CrossRef]
- Cord, A.F.; Meentemeyer, R.K.; Leitão, P.J.; Václavík, T. Modelling species distributions with remote sensing data: bridging disciplinary perspectives. J. Biogeogr. 2013, 40, 2226–2227. [Google Scholar] [CrossRef] [Green Version]
- Cord, A.F.; Klein, D.; Gernandt, D.S.; de la Rosa, J.A.P.; Dech, S. Remote sensing data can improve predictions of species richness by stacked species distribution models: A case study for mexican pines. J. Biogeogr. 2014, 41, 736–748. [Google Scholar] [CrossRef]
- Alcaraz, D.; Paruelo, J.; Cabello, J. Identification of current ecosystem functional types in the iberian peninsula. Glob. Ecol. Biogeogr. 2006, 15, 200–212. [Google Scholar] [CrossRef]
- Hogrefe, K.R.; Patil, V.P.; Ruthrauff, D.R.; Meixell, B.W.; Budde, M.E.; Hupp, J.W.; Ward, D.H. Normalized difference vegetation index as an estimator for abundance and quality of avian herbivore forage in Arctic Alaska. Remote Sens. 2017, 9, 1234. [Google Scholar] [CrossRef]
- Ames, G.M.; Wall, W.A.; Hohmann, M.G.; Wright, J.P. Trait space of rare plants in a fire-dependent ecosystem. Conserv. Biol. 2017, 31, 903–911. [Google Scholar] [CrossRef]
- Pausas, J.G.; Keeley, J.E. Wildfires as an Ecosystem Service. Front. Ecol. Environ. 2019, 17, 289–295. [Google Scholar] [CrossRef]
- Renwick, A.R.; Massimino, D.; Newson, S.E.; Chamberlain, D.E.; Pearce-Higgins, J.W.; Johnston, A. Modelling changes in species’ abundance in response to projected climate change. Divers. Distrib. 2012, 18, 121–132. [Google Scholar] [CrossRef]





| Competing Model | Predictors | LogLik | AICc | ΔAIC | wi | Explained Deviance | Spearman Correlation | MASE |
|---|---|---|---|---|---|---|---|---|
| EFAs + DEM | - EVImin - EVIdmic - SLO | −239.68 | 487.95 | 0.00 | 0.79 | 0.33 | 0.44 | 0.73 |
| EFAs + CLI | - EVImin - EVIdmic - BIO5 | −240.36 | 490.71 | 2.14 | 0.19 | 0.32 | 0.43 | 0.75 |
| EFAs | - EVImin - EVImn - EVIdmic | −240.87 | 492.85 | 2.76 | 0.20 | 0.31 | 0.43 | 0.77 |
| DEM | - ELE - SLO - ASP | −243.89 | 498.52 | 8.43 | 0.01 | 0.24 | 0.34 | 0.90 |
| CLI | - BIO5 - BIO15 - BIO19 | −247.62 | 505.97 | 15.88 | 0.00 | 0.15 | 0.31 | 0.96 |
| LCC | - DeFo - OShr - RoAr | −248.77 | 508.28 | 18.19 | 0.00 | 0.12 | 0.09 | 1 |
| Null model | - | −253.38 | 510.97 | 20.88 | 0.00 | 0.00 | - | 1 |
| Variable | OLS | 25th Quantile | 50th Quantile | 75th Quantile |
|---|---|---|---|---|
| Intercept | 22.95 (0.04) *** (489.98) | 10.95 (0.02) *** (379.31) | 18.07 (0.03) *** (454.55) | 28.88 (0.06) *** (438.24) |
| PpT | 1.66 (0.05) *** (32.80) | 1.11 (0.03) *** (36.50) | 1.50 (0.04) *** (37.26) | 2.61 (0.06) *** (38.30) |
| PpTa | 1.57 (0.04) *** (32.37) | 1.14 (0.03) *** (39.29) | 1.27 (0.04) *** (31.76) | 1.95 (0.06) *** (31.14) |
| Tmin | −0.65 (0.05) *** (−12.68) | −1.28 (0.02) *** (−44.71) | −1.30 (0.03) *** (−32.79) | −1.55 (0.06) *** (−24.24) |
| Tmina | −0.79 (0.04) *** (−16.23) | −0.39 (0.03) *** (−13.14) | −0.16 (0.03) *** (−4.23) | −0.46 (0.06) *** (−6.94) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arenas-Castro, S.; Regos, A.; Gonçalves, J.F.; Alcaraz-Segura, D.; Honrado, J. Remotely Sensed Variables of Ecosystem Functioning Support Robust Predictions of Abundance Patterns for Rare Species. Remote Sens. 2019, 11, 2086. https://0-doi-org.brum.beds.ac.uk/10.3390/rs11182086
Arenas-Castro S, Regos A, Gonçalves JF, Alcaraz-Segura D, Honrado J. Remotely Sensed Variables of Ecosystem Functioning Support Robust Predictions of Abundance Patterns for Rare Species. Remote Sensing. 2019; 11(18):2086. https://0-doi-org.brum.beds.ac.uk/10.3390/rs11182086
Chicago/Turabian StyleArenas-Castro, Salvador, Adrián Regos, João F. Gonçalves, Domingo Alcaraz-Segura, and João Honrado. 2019. "Remotely Sensed Variables of Ecosystem Functioning Support Robust Predictions of Abundance Patterns for Rare Species" Remote Sensing 11, no. 18: 2086. https://0-doi-org.brum.beds.ac.uk/10.3390/rs11182086