A Novel Approach for Estimation of Above-Ground Biomass of Sugar Beet Based on Wavelength Selection and Optimized Support Vector Machine
Abstract
:1. Introduction
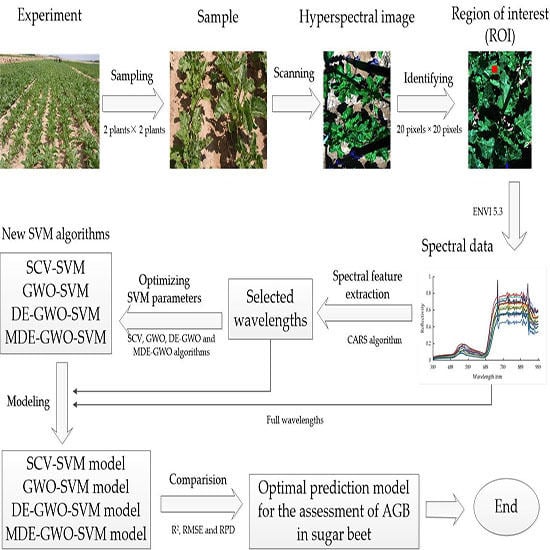
2. Materials and Methods
2.1. Experimental Design and Crop Growing
2.2. Measurements
2.2.1. Hyperspectral Images Measurement
2.2.2. Above-Ground Biomass (AGB) Measurement
2.3. Data Analysis and Modeling
2.3.1. Competitive Adaptive Reweighted Sampling Algorithm (CARS)
2.3.2. Grey Wolf Optimization Algorithm (GWO)
2.3.3. Differential Evolution Algorithm (DE)
2.3.4. Differential Evolution Grey Wolf Optimization Algorithm (DE–GWO)
2.3.5. Modified Differential Evolution Grey Wolf Optimization Algorithm (MDE–GWO)
2.3.6. Support Vector Machines Algorithm (SVM)
3. Results
3.1. Above-Ground Biomass (AGB) Variability
3.2. Correlation between Above-Ground Biomass (AGB) and Canopy Reflectance Wavelength
3.3. Characteristic Wavelengths Selection with Competitive Adaptive Reweighted Sampling (CARS)
3.4. Modified Differential Evolution Grey Wolf Optimization (MDE–GWO)
3.5. Support Vector Machine (SVM) Models for Above-Ground Biomass (AGB) Prediction
4. Discussion
4.1. Important Waveband for the Prediction of Above-Ground Biomass (AGB)
4.2. Performance of Support Vector Machine (SVM) Models
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Yue, J.B.; Yang, G.J.; Li, C.C.; Li, Z.H.; Wang, Y.J.; Feng, H.K.; Xu, B. Estimation of winter wheat above-ground biomass using unmanned aerial vehicle-based snapshot hyperspectral sensor and crop height improved models. Remote Sens. 2017, 9, 708. [Google Scholar] [CrossRef] [Green Version]
- Yue, J.B.; Yang, G.J.; Feng, H.K. Comparative of remote sensing estimation models of winter wheat biomass based on random forest algorithm. Trans. Chin. Soc. Agric. Eng. 2016, 32, 175–182. [Google Scholar]
- Hensgen, F.; Bühle, L.; Wachendorf, M. The effect of harvest, mulching and low-dose fertilization of liquid digestate on above ground biomass yield and diversity of lower mountain semi-natural grasslands. Agric. Ecosyst. Environ. 2016, 216, 283–292. [Google Scholar] [CrossRef]
- Martin, M.E.; Plourde, L.C.; Ollinger, S.V.; Smith, M.L.; McNeil, B.E. A generalizable method for remote sensing of canopy nitrogen across a wide range of forest ecosystems. Remote Sens. Environ. 2008, 112, 3511–3519. [Google Scholar] [CrossRef]
- Nguyen, H.T.; Lee, B.W. Assessment of rice leaf growth and nitrogen status by hyperspectral canopy reflectance and partial least square regression. Eur. J. Agron. 2006, 24, 349–356. [Google Scholar] [CrossRef]
- Thenkabail, P.S.; Mariotto, I.; Gumma, M.K.; Middleton, E.M.; Landis, D.R.; Huemmrich, K.F. Selection of hyperspectral narrowbands (hnbs) and composition of hyperspectral twoband vegetation indices (HVIS) for biophysical characterization and discrimination of crop types using field reflectance and hyperion/EO-1 data. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2013, 6, 427–439. [Google Scholar] [CrossRef] [Green Version]
- Sellami, A.; Farah, M.; Farah, I.R.; Solaiman, B. Hyperspectral imagery classification based on semi-supervised 3-D deep neural network and adaptive band selection. Expert Syst. Appl. 2019, 129, 246–259. [Google Scholar] [CrossRef]
- Wang, M.W.; Wu, C.M.; Wang, L.Z.; Xiang, D.X.; Huang, X.H. A feature selection approach for hyperspectral image based onmodified ant lion optimizer. Knowl. Based Syst. 2019, 168, 39–48. [Google Scholar] [CrossRef]
- Yang, B.H.; Chen, J.L.; Chen, L.H.; Cao, W.X.; Yao, X.; Zhu, Y. Estimation model of wheat canopy nitrogen content based on sensitive bands. Trans. Chin. Soc. Agric. Eng. 2015, 31, 176–182. [Google Scholar]
- Xiao, H.; Li, A.; Li, M.Y.; Sun, Y.; Tu, K.; Wang, S.J.; Pan, L.Q. Quality assessment and discrimination of intact white and red grapes from Vitis vinifera L. at five ripening stages by visible and near-infrared spectroscopy. Sci. Hortic. 2018, 233, 99–107. [Google Scholar] [CrossRef]
- Hansen, P.M.; Schjoerring, J.K. Reflectance measurement of canopy biomass and nitrogen status in wheat crops using normalized difference vegetation indices and partial least squares regression. Remote Sens. Environ. 2003, 86, 542–553. [Google Scholar] [CrossRef]
- Karimi, Y.; Prasher, S.O.; Patel, R.M.; Kim, S.H. Application of support vector machine technology for weed and nitrogen stress detection in corn. Comput. Electron. Agric. 2005, 51, 99–109. [Google Scholar] [CrossRef]
- Clevers, J.G.P.W.; van der Heijden, G.W.A.M.; Verzakov, S.; Schaepman, M.E. Estimating grassland biomass using SVM band shaving of hyperspectral data. Photogramm. Eng. Remote Sens. 2007, 73, 1141–1148. [Google Scholar] [CrossRef] [Green Version]
- Yuan, H.H.; Yang, G.J.; Li, C.C.; Wang, Y.J.; Liu, J.G.; Yu, H.Y.; Feng, H.K.; Xu, B.; Zhao, X.Q.; Yang, X.D. Retrieving soybean leaf area index from unmanned aerial vehicle hyperspectral remote sensing: Analysis of RF, ANN, and SVM regression models. Remote Sens. 2017, 9, 309. [Google Scholar] [CrossRef] [Green Version]
- Tarabalka, Y.; Fauvel, M.; Chanussot, J.; Benediktsson, J.A. SVM- and MRF-Based Method for Accurate Classification of Hyperspectral Images. IEEE Geosci. Remote Sens. Lett. 2010, 7, 736–740. [Google Scholar] [CrossRef] [Green Version]
- Kuo, B.C.; Ho, H.H.; Li, C.H.; Hung, C.C.; Taur, J.S. A kernel-based feature selection method for SVM with RBF kernel for hyperspectral image classification. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2014, 7, 317–326. [Google Scholar]
- Zhang, S.; Zhou, Y.Q.; Li, Z.M.; Pan, W. Grey wolf optimizer for unmanned combat aerial vehicle path planning. Adv. Eng. Softw. 2016, 99, 121–136. [Google Scholar] [CrossRef] [Green Version]
- Khairuzzaman, A.K.M.; Chaudhury, S. Multilevel thresholding using grey wolf optimizer for image segmentation. Expert Syst. Appl. 2017, 86, 64–76. [Google Scholar] [CrossRef]
- Yamany, W.; Emary, E.; Hassanien, A.E. New rough set attribute reduction algorithm based on grey wolf optimization. In Advances in Intelligent Systems and Computing; Springer: Cham, Switzerland, 2016; pp. 241–251. [Google Scholar]
- Wang, X.F.; Zhao, H.; Han, T.; Zhou, H.; Li, C. A grey wolf optimizer using Gaussian estimation of distribution and its application in the multi-UAV multi-target urban tracking problem. Appl. Soft Comput. J. 2019, 78, 240–260. [Google Scholar] [CrossRef]
- Zhu, A.J.; Xu, C.P.; Li, Z.; Wu, J.; Liu, Z.B. Hybridizing grey wolf optimization with differential evolution for global optimization and test scheduling for 3D stacked SoC. J. Syst. Eng. Electron. 2015, 26, 317–328. [Google Scholar] [CrossRef]
- Debnath, M.K.; Mallick, R.K.; Sahu, B.K. Application of Hybrid Differential Evolution–Grey Wolf Optimization Algorithm for Automatic Generation Control of a Multi-Source Interconnected Power System Using Optimal Fuzzy–PID Controller. Electr. Power Compon. Syst. 2017, 45, 2104–2117. [Google Scholar] [CrossRef]
- Xu, S.J.; Long, W. Improved grey wolf optimizer algorithm based on stochastic convergence factor and differential mutation. Sci. Technol. Eng. 2018, 18, 252–256. [Google Scholar]
- Wang, M.; Tang, M.Z. Novel grey wolf optimization algorithm based on nonlinear convergence factor. Appl. Res. Comput. 2016, 33, 3648–3653. [Google Scholar]
- Tian, X.; Li, J.B.; Wang, Q.Y.; Fan, S.X.; Huang, W.Q.; Zhao, C.J. A multi-region combined model for non-destructive prediction of soluble solids content in apple, based on brightness grade segmentation of hyperspectral imaging. Biosyst. Eng. 2019, 183, 110–120. [Google Scholar] [CrossRef]
- Ariana, D.P.; Lu, R. Hyperspectral waveband selection for internal defect detection of pickling cucumbers and whole pickles. Comput. Electron. Agric. 2010, 74, 137–144. [Google Scholar] [CrossRef]
- Li, H.D.; Liang, Y.Z.; Xu, Q.S.; Cao, D.S. Key wavelengths screening using competitive adaptive reweighted sampling method for multivariate calibration. Anal. Chim. Acta 2009, 648, 77–84. [Google Scholar] [CrossRef]
- He, H.J.; Sun, D.W.; Wu, D. Rapid and real-time prediction of lactic acid bacteria (LAB) in farmed salmon flesh using near-infrared (NIR) hyperspectral imaging combined with chemometric analysis. Food Res. Int. 2014, 62, 476–483. [Google Scholar] [CrossRef]
- Mirjalili, S.; Mirjalili, S.M.; Lewis, A. Grey Wolf Optimizer. Adv. Eng. Softw. 2014, 69, 46–61. [Google Scholar] [CrossRef] [Green Version]
- Sharma, P.; Sundaram, S.; Sharma, M.; Sharma, A.; Gupta, D. Diagnosis of Parkinson’s disease using modified grey wolf optimization. Cogn. Syst. Res. 2019, 54, 100–115. [Google Scholar] [CrossRef]
- Storn, R.; Price, K. Differential Evolution-A Simple and Efficient Adaptive Scheme for Global Optimization over Continuous Spaces; Technical Report TR-95-012; International Computer Science Institute: Berkley, CA, USA, 1995; Volume 23. [Google Scholar]
- Sayah, S.; Zehar, K. Modified differential evolution algorithm for optimal power flow with non-smooth cost functions. Energy Convers. Manag. 2008, 49, 3036–3042. [Google Scholar] [CrossRef]
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Vapnik, V.N. The Nature of Statistical Learning Theory, 2nd ed.; Springer: New York, NY, USA, 2000. [Google Scholar]
- Todorova, M.; Mouazen, A.M.; Lange, H.; Atanassova, S. Potential of near-infrared spectroscopy for measurement of heavy metals in soil as affected by calibration set size. Water Air Soil Pollut. 2014, 225, 2036. [Google Scholar] [CrossRef]
- Cheng, W.W.; Sun, D.W.; Pu, H.B.; Wei, Q.Y. Characterization of myofibrils cold structural deformation degrees of frozen pork using hyperspectral imaging coupled with spectral angle mapping algorithm. Food Chem. 2018, 239, 1001–1008. [Google Scholar] [CrossRef]
- Brodersen, C.R.; Vogelmann, T.C. Do changes in light direction affect absorption profiles in leaves? Funct. Plant Biol. 2010, 37, 403–412. [Google Scholar] [CrossRef]
- Inoue, Y.; Moran, M.S.; Horie, T. Analysis of spectral measurements in paddy field for predicting rice growth and yield based on simple crop simulation model. Plant Prod. Sci. 1998, 1, 269–279. [Google Scholar] [CrossRef]
- Clevers, J.G.P.W.; Kooistra, L.; Schaepman, M.E. Estimating canopy water content using hyperspectral remote sensing data. Int. J. Appl. Earth Obs. Geoinf. 2010, 2, 119–125. [Google Scholar] [CrossRef]
- Thenkabail, P.S.; Lyon, J.G.; Huete, A. Advance in hyperspectral remote sensing of vegetation of vegetation and agricultural croplands. In Hyperspectral Remote Sensing of Vegetation; Thenkabail, P.S., Lyon, J.G., Huete, A., Eds.; CRC Press: Boca Raton, FL, USA, 2011; pp. 3–36. [Google Scholar]
- Liu, H.; Fu, Y.M.; Hu, D.W.; Yu, J.; Liu, H. Effect of green, yellow and purple radiation on biomass, photosynthesis, morphology and soluble sugar content of leafy lettuce via spectral wavebands “knock out”. Sci. Hortic. 2018, 236, 10–17. [Google Scholar] [CrossRef]
- Sun, H.; Li, M.Z.; Zhao, Y.; Zhang, Y.E.; Wang, X.M.; Li, X.H. The spectral characteristics and chlorophyll content at winter wheat growth stages. Spectrosc. Spectr. Anal. 2010, 30, 192–196. [Google Scholar]
- Preece, J.E.; Read, P.E. The Biology of Horticulture: An Introductory Textbook; John Wiley: New York, NY, USA, 1993. [Google Scholar]
- Clark, M.L.; Roberts, D.A.; Ewel, J.J.; Clark, D.B. Estimation of tropical rain forest aboveground biomass with small-footprint lidar and hyperspectral sensors. Remote Sens. Environ. 2011, 115, 231–2942. [Google Scholar] [CrossRef]
- Gnyp, M.L.; Bareth, G.; Li, F.; Lenz-Wiedemann, V.I.S.; Koppe, W.; Miao, Y.X.; Henning, S.D.; Jia, L.L.; Laudien, R.; Chen, X.P.; et al. Development and implementation of a multiscale biomass model using hyperspectral vegetation indices for winter wheat in the North China Plain. Int. J. Appl. Earth Obs. Geoinf. 2014, 33, 232–242. [Google Scholar] [CrossRef]
- Chang, C.W.; Laird, D.; Mausbach, M.J.; Hurburgh, C.R. Near infrared reflectance spectroscopy: Principal components regression analysis of soil properties. Agric. Biosyst. Eng. 2001, 3, 480–490. [Google Scholar] [CrossRef] [Green Version]
- Mouazen, A.M.; Baerdemaeker, J.D.; Ramon, H. Effect of wavelength range on the measurement accuracy of some selected soil constituents using visual-near infrared spectroscopy. J. Near Infrared Spectrosc. 2006, 14, 189–199. [Google Scholar] [CrossRef]
- Li, L.T.; Wang, S.Q.; Ren, T.; Wei, Q.Q.; Ming, J.; Li, J.; Li, X.K.; Cong, R.H.; Lu, J.W. Ability of models with effective wavelengths to monitor nitrogen and phosphorus status of winter oilseed rape leaves using in situ canopy spectroscopy. Field Crop. Res. 2018, 215, 173–186. [Google Scholar] [CrossRef]
- Jin, H.L.; Favaroy, P.; Soatto, S. Real-time feature tracking and outlier rejection with changes in illumination. In Proceedings of the IEEE Eighth International Conference on Computer Vision, Vancouver, BC, Canada, 7–14 July 2001; Volume 1, pp. 684–689. [Google Scholar]
- Li, N.; Zhu, X.F.; Pan, Y.Z.; Zhan, P. Optimized SVM based on artificial bee colony algorithm for remote sensing image classification. J. Remote Sens. 2018, 22, 559–569. [Google Scholar]










| Year | Area (m2) | Cultivars | Soil Properties | Planting Pattern | N Rates (kg/hm2) | Soil Texture |
|---|---|---|---|---|---|---|
| 2014 | 4200 | KWS1676 | Organic C: 13.04 g/kg Total N: 0.76 g/kg Available P: 12.48 mg/kg Available K: 114.2 mg/kg pH: 8.2 | Transplant | 0 (N0) 15 (N1) 32 (N2) 76 (N3) 108 (N4) 163 (N5) 217 (N6) | Sandy loam |
| 2015 | 1800 | KWS9147 | Organic C: 23.6 g/kg Total N: 1.46 g/kg Available P: 42 mg/kg Available K: 156 mg/kg pH: 7.3 | Direct seeding | 0 (N0) 80 (N1) 120 (N2) 200 (N3) | Loam |
| 2018 | 1200 | KWS1231 | Organic C: 16.32 g/kg Total N: 0.78 g/kg Available P: 37.71 mg/kg Available K: 314.6 mg/kg pH: 8.6 | Direct seeding | 0 (N0) 70 (N1) 90 (N2) 116 (N3) 130 (N4) 150 (N5) | Clay |
| Growth Stage | 2014 | 2015 | 2018 | |||
|---|---|---|---|---|---|---|
| Measurement Date | Number of Samples | Measurement Date | Number of Samples | Measurement Date | Number of Samples | |
| Rapid growth stage of leaf cluster | 23 June and 10 July | 28 | 8 July and 20 July | 12 | 27 June and 14 July | 24 |
| Sugar growth stage | 25 July and 17 August | 28 | 13 August and 20 August | 12 | 29 July and 9 August | 24 |
| Sugar accumulation stage | 30 August and 15 September | 28 | 31 August and 15 September | 12 | 26 August and 15 September | 24 |
| Stage | Calibration Set | Validation Set | ||||
|---|---|---|---|---|---|---|
| 2014 | 2015 | 2018 | 2014 | 2015 | 2018 | |
| Three studied growth stages | 28 | 12 | 24 | 28 | / | / |
| / | 12 | / | ||||
| / | / | 24 | ||||
| Date Set | Summary Statistics | Rapid Growth Stage of Leaf Cluster | Sugar Growth Stage | Sugar Accumulation Stage | |
|---|---|---|---|---|---|
| Calibration set (n = 64) | Mean | 344.77 | 530.25 | 470.81 | |
| SD a | 156.61 | 190.12 | 164.34 | ||
| Max | 596.95 | 1012.04 | 789.77 | ||
| Min | 33.47 | 82.64 | 138.54 | ||
| Validation set | 2014 (n = 28) | Mean | 425.13 | 541.87 | 542.94 |
| SD | 116.12 | 51.67 | 155.55 | ||
| Max | 556.99 | 666.33 | 792.21 | ||
| Min | 86.11 | 444.32 | 177.23 | ||
| 2015 (n = 12) | Mean | 133.30 | 400.34 | 332.41 | |
| SD | 67.68 | 65.95 | 41.61 | ||
| Max | 269.84 | 493.32 | 394.73 | ||
| Min | 49.41 | 233.35 | 256.70 | ||
| 2018 (n = 24) | Mean | 443.88 | 570.03 | 598.79 | |
| SD | 103.61 | 57.16 | 63.69 | ||
| Max | 559.06 | 651.93 | 685.77 | ||
| Min | 98.54 | 427.94 | 452.61 | ||
| Growth Stages | Inputs | Model | Calibration Set | Validation Set | ||||
|---|---|---|---|---|---|---|---|---|
| R2 a | RMSE b (g/m2) | Year | R2 | RMSE (g/m2) | RPD c | |||
| Rapid Growth Stage of Leaf Cluster | All Bands | GWO | 0.76 | 79.69 | 2014 | 0.61 | 82.82 | 0.99 |
| 2015 | 0.60 | 59.70 | 0.77 | |||||
| 2018 | 0.70 | 71.79 | 0.81 | |||||
| DE–GWO | 0.82 | 113.16 | 2014 | 0.80 | 58.54 | 1.52 | ||
| 2015 | 0.61 | 53.30 | 1.16 | |||||
| 2018 | 0.70 | 70.65 | 0.82 | |||||
| MDE–GWO | 0.86 | 61.41 | 2014 | 0.84 | 59.26 | 1.97 | ||
| 2015 | 0.64 | 49.73 | 1.21 | |||||
| 2018 | 0.75 | 72.98 | 0.90 | |||||
| CARS | GWO | 0.78 | 76.84 | 2014 | 0.71 | 84.25 | 1.35 | |
| 2015 | 0.64 | 42.83 | 1.27 | |||||
| 2018 | 0.70 | 80.26 | 1.25 | |||||
| DE–GWO | 0.82 | 70.73 | 2014 | 0.77 | 70.89 | 1.64 | ||
| 2015 | 0.68 | 46.21 | 1.36 | |||||
| 2018 | 0.74 | 67.01 | 1.26 | |||||
| MDE–GWO | 0.84 | 67.54 | 2014 | 0.80 | 53.69 | 1.97 | ||
| 2015 | 0.74 | 46.17 | 1.42 | |||||
| 2018 | 0.75 | 65.68 | 1.71 | |||||
| Sugar Growth Stage | All Bands | GWO | 0.78 | 119.66 | 2014 | 0.69 | 37.56 | 1.11 |
| 2015 | 0.52 | 70.47 | 1.15 | |||||
| 2018 | 0.49 | 47.53 | 0.96 | |||||
| DE–GWO | 0.82 | 116.94 | 2014 | 0.71 | 30.92 | 1.33 | ||
| 2015 | 0.58 | 67.81 | 1.17 | |||||
| 2018 | 0.52 | 40.06 | 1.03 | |||||
| MDE–GWO | 0.89 | 81.27 | 2014 | 0.82 | 27.66 | 2.01 | ||
| 2015 | 0.69 | 47.13 | 1.21 | |||||
| 2018 | 0.69 | 35.60 | 1.40 | |||||
| CARS | GWO | 0.80 | 156.81 | 2014 | 0.75 | 39.68 | 1.47 | |
| 2015 | 0.65 | 52.55 | 1.28 | |||||
| 2018 | 0.74 | 46.29 | 1.25 | |||||
| DE–GWO | 0.82 | 154.77 | 2014 | 0.77 | 32.13 | 1.60 | ||
| 2015 | 0.72 | 57.46 | 1.38 | |||||
| 2018 | 0.78 | 66.58 | 0.27 | |||||
| MDE–GWO | 0.85 | 154.43 | 2014 | 0.80 | 30.16 | 2.03 | ||
| 2015 | 0.78 | 32.35 | 1.97 | |||||
| 2018 | 0.80 | 37.03 | 1.69 | |||||
| Sugar Accumulation Stage | All Bands | GWO | 0.75 | 89.07 | 2014 | 0.70 | 99.75 | 1.14 |
| 2015 | 0.42 | 34.44 | 0.56 | |||||
| 2018 | 0.70 | 40.14 | 1.10 | |||||
| DE–GWO | 0.81 | 77.13 | 2014 | 0.69 | 94.10 | 1.31 | ||
| 2015 | 0.58 | 36.87 | 0.92 | |||||
| 2018 | 0.71 | 34.89 | 1.60 | |||||
| MDE–GWO | 0.83 | 69.83 | 2014 | 0.74 | 81.86 | 1.61 | ||
| 2015 | 0.61 | 27.24 | 1.32 | |||||
| 2018 | 0.74 | 32.52 | 1.65 | |||||
| CARS | GWO | 0.80 | 78.87 | 2014 | 0.71 | 101.45 | 1.12 | |
| 2015 | 0.65 | 25.78 | 1.48 | |||||
| 2018 | 0.69 | 40.04 | 1.67 | |||||
| DE–GWO | 0.82 | 72.27 | 2014 | 0.70 | 87.89 | 1.71 | ||
| 2015 | 0.67 | 30.05 | 1.56 | |||||
| 2018 | 0.72 | 36.75 | 1.70 | |||||
| MDE–GWO | 0.83 | 72.00 | 2014 | 0.73 | 104.08 | 1.72 | ||
| 2015 | 0.69 | 40.77 | 1.61 | |||||
| 2018 | 0.74 | 40.17 | 1.95 | |||||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Tian, H.; Wang, D.; Li, H.; Mouazen, A.M. A Novel Approach for Estimation of Above-Ground Biomass of Sugar Beet Based on Wavelength Selection and Optimized Support Vector Machine. Remote Sens. 2020, 12, 620. https://0-doi-org.brum.beds.ac.uk/10.3390/rs12040620
Zhang J, Tian H, Wang D, Li H, Mouazen AM. A Novel Approach for Estimation of Above-Ground Biomass of Sugar Beet Based on Wavelength Selection and Optimized Support Vector Machine. Remote Sensing. 2020; 12(4):620. https://0-doi-org.brum.beds.ac.uk/10.3390/rs12040620
Chicago/Turabian StyleZhang, Jing, Haiqing Tian, Di Wang, Haijun Li, and Abdul Mounem Mouazen. 2020. "A Novel Approach for Estimation of Above-Ground Biomass of Sugar Beet Based on Wavelength Selection and Optimized Support Vector Machine" Remote Sensing 12, no. 4: 620. https://0-doi-org.brum.beds.ac.uk/10.3390/rs12040620