Towards Tree Green Crown Volume: A Methodological Approach Using Terrestrial Laser Scanning
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Area
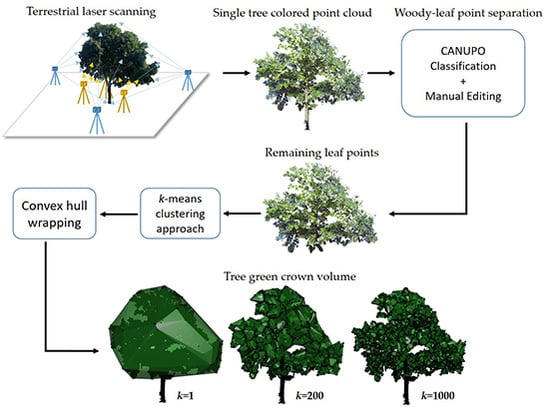
2.2. Workflow Outline
2.3. Definition
2.4. Field Measurements
2.5. Terrestrial Laser Scanning
2.6. Determining of Tree Green Crown Volume
2.7. The Tree Green Crown Volume Index
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Von Oheimb, G.; Lang, A.C.; Bruelheide, H.; Forrester, D.I.; Wäsche, I.; Yu, M.; Härdtle, W. Individual-tree radial growth in a subtropical broad-leaved forest: The role of local neighbourhood competition. For. Ecol. Manag. 2011, 261, 499–507. [Google Scholar] [CrossRef]
- Krajicek, J.; Brinkman, K.; Gingrich, S. Crown Competition—A Measure of Density. For. Sci. 1961, 7, 35–42. [Google Scholar] [CrossRef]
- Schuldt, A.; Ebeling, A.; Kunz, M.; Staab, M.; Guimarães-Steinicke, C.; Bachmann, D.; Buchmann, N.; Durka, W.; Fichtner, A.; Fornoff, F.; et al. Multiple plant diversity components drive consumer communities across ecosystems. Nat. Commun. 2019, 10, 1460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nowak, D.J.; Rowntree, R.A.; McPherson, E.G.; Sisinni, S.M.; Kerkmann, E.R.; Stevens, J.C. Measuring and analyzing urban tree cover. Landsc. Urban Plan. 1996, 36, 49–57. [Google Scholar] [CrossRef] [Green Version]
- FAO. Global Forest Resources Assessment 2020; Terms and Definitions FRA 2020; Food and Agriculture Organization: Rome, Italy, 2018; p. 26. [Google Scholar]
- Zeide, B. Fractal analysis of foliage distribution in loblolly pine crowns. Can. J. For. Res. 1998, 28, 106–114. [Google Scholar] [CrossRef]
- Javacheff, C.V.; De Guillebon, J.C.D. Wrapped Trees; Fondation Beyeler and Berower Park: Riehen, Switzerland, 1997–1998; Available online: https://christojeanneclaude.net/mobile/projects?p=wrapped-trees (accessed on 26 May 2020).
- Bréda, N.J.J. Leaf area index. In Encyclopedia of Ecology; Elsevier: New York, NY, USA, 2018; ISBN 9780444641304. [Google Scholar]
- Soma, M.; Pimont, F.; Durrieu, S.; Dupuy, J.L. Enhanced measurements of leaf area density with T-LiDAR: Evaluating and calibrating the effects of vegetation heterogeneity and scanner properties. Remote Sens. 2018, 10, 1580. [Google Scholar] [CrossRef] [Green Version]
- Béland, M.; Baldocchi, D.D.; Widlowski, J.L.; Fournier, R.A.; Verstraete, M.M. On seeing the wood from the leaves and the role of voxel size in determining leaf area distribution of forests with terrestrial LiDAR. Agric. For. Meteorol. 2014, 184, 82–97. [Google Scholar] [CrossRef]
- Pimont, F.; Allard, D.; Soma, M.; Dupuy, J.L. Estimators and confidence intervals for plant area density at voxel scale with T-LiDAR. Remote Sens. Environ. 2018, 215, 343–370. [Google Scholar] [CrossRef]
- Wang, D.; Song, H.; Tie, Z.; Zhang, W.; He, D. Recognition and localization of occluded apples using K-means clustering algorithm and convex hull theory: A comparison. Multimed. Tools Appl. 2016, 75, 3177–3198. [Google Scholar] [CrossRef]
- Rusu, R.B.; Cousins, S. 3D is here: Point Cloud Library (PCL). In Proceedings of the 2011 IEEE International Conference on Robotics and Automation, Shanghai, China, 9–13 May 2011. [Google Scholar]
- Brodu, N.; Lague, D. 3D terrestrial lidar data classification of complex natural scenes using a multiscale dimensionality criterion: Applications in geomorphology. ISPRS J. Photogramm. Remote Sens. 2012, 68, 121–134. [Google Scholar] [CrossRef] [Green Version]
- MacQueen, J. Some methods for classification and analysis of multivariate observations. In Proceedings of the Fifth Berkeley Symposium on Mathematical Statistics and Probability; University of California Press: Berkeley, CA, USA, 1967. [Google Scholar]
- Schlager, S. Morpho and Rvcg—Shape Analysis in R. In Statistical Shape and Deformation Analysis; Elsevier: New York, NY, USA, 2017. [Google Scholar]
- Silva, C.A.; Crookston, N.L.; Hudak, A.T.; Vierling, L.A. rLiDAR: An R Package for Reading, Processing and Visualizing Lidar (Light Detection and Ranging) Data, Version 0.1.1. 2017. Available online: https://cran.r-project.org/web/packages/rLiDAR/index.html (accessed on 5 June 2020).
- Maltamo, M.; Næsset, E.; Vauhkonen, J. Forestry Applications of Airborne Laser Scanning: Concepts and Case Studies; Springer: Dordrecht, The Netherlands, 2014; ISBN 9789401786621. [Google Scholar]
- Wang, D.; Hollaus, M.; Pfeifer, N. Feasibility of machine learning methods for separating wood and leaf points from terrestrial laser scanning data. ISPRS Ann. Photogramm. Remote Sens. Spatial Inf. Sci. 2017, IV-2/W4, 157–164. [Google Scholar] [CrossRef] [Green Version]
- Vicari, M.B.; Disney, M.; Wilkes, P.; Burt, A.; Calders, K.; Woodgate, W. Leaf and wood classification framework for terrestrial LiDAR point clouds. Methods Ecol. Evol. 2019, 10, 680–694. [Google Scholar] [CrossRef] [Green Version]
- Kukunda, C.B.; Beckschäfer, P.; Magdon, P.; Schall, P.; Wirth, C.; Kleinn, C. Scale-guided mapping of forest stand structural heterogeneity from airborne LiDAR. Ecol. Indic. 2019, 102, 410–425. [Google Scholar] [CrossRef]
- Kleinn, C.; Kändler, G.; Schnell, S. Estimating forest edge length from forest inventory sample data. Can. J. For. Res. 2011, 41, 1–10. [Google Scholar] [CrossRef]







| Terms | Elements of the Definition | Notation Here |
|---|---|---|
| Crown base height | The vertical distance from ground to the lowest leaf layer of the tree crown meaning the first foliage advancing upwards from the ground. | CBH |
| Crown length | The vertical distance between crown base and crown (tree) top. | CL |
| Crown width | The width of horizontal tree crown projections in one direction. Here, we used two fixed directions and averaged the measurements. | CW |
| Tree height | The vertical distance from the ground level to the level of the crown (tree) top. | H |
| Diameter at breast height | The diameter of the stem at the height of 1.3m above ground measured perpendicularly to the stem axis. | DBH |
| Tree Id | Species | Location | DBH | H | CBH | Averaged CW |
|---|---|---|---|---|---|---|
| (cm) | (m) | (m) | (m) | |||
| Tree1 | Acer platanoides | Botanical Garden | 35.8 | 12.5 | 1.0 | 7.4 |
| Tree2 | Acer platanoides | Botanical Garden | 34.0 | 10.5 | 1.5 | 6.2 |
| Tree3 | Acer pseudoplatanus | Roadside | 58.0 | 17.1 | 0.7 | 11.6 |
| Tree4 | Acer pseudoplatanus | Roadside | 61.2 | 14.8 | 0.9 | 11.85 |
| Tree5 | Acer pseudoplatanus | Roadside | 46.6 | 12.8 | 0.8 | 12.45 |
| Tree6 | Acer pseudoplatanus | Roadside | 35.6 | 10.9 | 1.0 | 12.0 |
| Tree7 | Acer pseudoplatanus | Roadside | 43.8 | 17.4 | 1.6 | 13.7 |
| Tree8 | Acer pseudoplatanus | Roadside | 88.2 | 22 | 1.0 | 13.9 |
| Tree9 | Acer pseudoplatanus | Roadside | 49.5 | 10.7 | 1.0 | 13.65 |
| Tree10 | Acer pseudoplatanus | Roadside | 43.0 | 10.5 | 2.0 | 11.25 |
| Tree11 | Acer pseudoplatanus | Roadside | 16.9 | 14 | 1.8 | 6.5 |
| Tree12 | Acer pseudoplatanus | Roadside | 20.4 | 13.6 | 2.4 | 7.75 |
| Tree13 | Acer pseudoplatanus | Roadside | 55.8 | 15.3 | 2.6 | 14.25 |
| Tree14 | Acer pseudoplatanus | Roadside | 36.2 | 11.6 | 2.1 | 10.85 |
| Tree15 | Aesculus hippocastanum | Roadside | 72.8 | 18.2 | 2.2 | 14.3 |
| Tree16 | Aesculus × carnea | Botanical Garden | 20.0 | 6.3 | 1.0 | 6.35 |
| Tree17 | Carpinus betulus | Roadside | 53.6 | 14.6 | 0.5 | 11.5 |
| Tree18 | Fagus sylvatica | Roadside | 42.7 | 12.5 | 1.5 | 8.3 |
| Tree19 | Liriodendron tulipifera | Roadside | 39.5 | 16.8 | 0.0 | 10.35 |
| Tree20 | Prunus avium | Botanical Garden | 42.6 | 8.7 | 1.5 | 8.0 |
| Tree21 | Quercus robur | Roadside | 43.5 | 12.4 | 2.4 | 10.6 |
| Tree22 | Quercus robur | Roadside | 64.1 | 15.4 | 1.4 | 15.8 |
| Tree23 | Quercus robur | Roadside | 59.1 | 12.8 | 1.0 | 17.35 |
| Tree24 | Quercus robur | Roadside | 29.4 | 12.4 | 3.0 | 12.2 |
| Tree25 | Tilia cordata | Roadside | 43.5 | 15.0 | 2.0 | 11.15 |
| Tree26 | Tilia cordata | Roadside | 59.9 | 16.1 | 1.4 | 11.6 |
| Tree Id | Species | CVol convex hull (m3) | TGCVI When k = | ||
|---|---|---|---|---|---|
| 100 | 500 | 1000 | |||
| Tree1 | Acer platanoides | 289 | 0.63 | 0.41 | 0.32 |
| Tree2 | Acer platanoides | 196 | 0.63 | 0.37 | 0.29 |
| Tree3 | Acer pseudoplatanus | 1217 | 0.66 | 0.41 | 0.33 |
| Tree4 | Acer pseudoplatanus | 990 | 0.62 | 0.40 | 0.30 |
| Tree5 | Acer pseudoplatanus | 1026 | 0.66 | 0.44 | 0.36 |
| Tree6 | Acer pseudoplatanus | 700 | 0.58 | 0.38 | 0.30 |
| Tree7 | Acer pseudoplatanus | 1576 | 0.64 | 0.40 | 0.32 |
| Tree8 | Acer pseudoplatanus | 1736 | 0.66 | 0.43 | 0.35 |
| Tree9 | Acer pseudoplatanus | 812 | 0.52 | 0.26 | 0.18 |
| Tree10 | Acer pseudoplatanus | 473 | 0.38 | 0.20 | 0.14 |
| Tree11 | Acer pseudoplatanus | 163 | 0.47 | 0.28 | 0.22 |
| Tree12 | Acer pseudoplatanus | 270 | 0.41 | 0.23 | 0.17 |
| Tree13 | Acer pseudoplatanus | 1234 | 0.56 | 0.39 | 0.31 |
| Tree14 | Acer pseudoplatanus | 681 | 0.65 | 0.40 | 0.31 |
| Tree15 | Aesculus hippocastanum | 1814 | 0.69 | 0.48 | 0.39 |
| Tree16 | Aesculus × carnea | 111 | 0.58 | 0.32 | 0.22 |
| Tree17 | Carpinus betulus | 817 | 0.55 | 0.37 | 0.28 |
| Tree18 | Fagus sylvatica | 380 | 0.64 | 0.43 | 0.34 |
| Tree19 | Liriodendron tulipifera | 1049 | 0.57 | 0.38 | 0.30 |
| Tree20 | Prunus avium | 347 | 0.55 | 0.29 | 0.21 |
| Tree21 | Quercus robur | 620 | 0.45 | 0.23 | 0.18 |
| Tree22 | Quercus robur | 2092 | 0.54 | 0.28 | 0.20 |
| Tree23 | Quercus robur | 1729 | 0.54 | 0.28 | 0.20 |
| Tree24 | Quercus robur | 730 | 0.49 | 0.28 | 0.19 |
| Tree25 | Tilia cordata | 904 | 0.61 | 0.36 | 0.28 |
| Tree26 | Tilia cordata | 1003 | 0.68 | 0.51 | 0.42 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, Z.; Kleinn, C.; Nölke, N. Towards Tree Green Crown Volume: A Methodological Approach Using Terrestrial Laser Scanning. Remote Sens. 2020, 12, 1841. https://0-doi-org.brum.beds.ac.uk/10.3390/rs12111841
Zhu Z, Kleinn C, Nölke N. Towards Tree Green Crown Volume: A Methodological Approach Using Terrestrial Laser Scanning. Remote Sensing. 2020; 12(11):1841. https://0-doi-org.brum.beds.ac.uk/10.3390/rs12111841
Chicago/Turabian StyleZhu, Zihui, Christoph Kleinn, and Nils Nölke. 2020. "Towards Tree Green Crown Volume: A Methodological Approach Using Terrestrial Laser Scanning" Remote Sensing 12, no. 11: 1841. https://0-doi-org.brum.beds.ac.uk/10.3390/rs12111841