Applications of Graphene Quantum Dots in Biomedical Sensors
Abstract
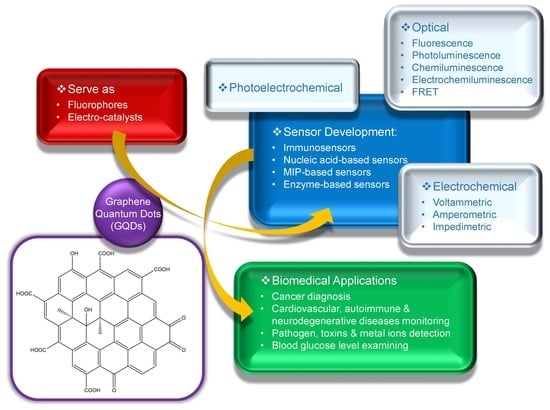
:1. Introduction
1.1. History, Definition and Classification of Biosensors
1.2. Role of Nanomaterials in Biosensing
1.3. Ideal Properties of Graphene Quantum Dots (GQDs)
1.4. Approaches to Synthesize GQDs for Biomedical Sensors
- (a)
- (b)
2. Optical GQD Sensors in Biomedical Diagnostics
2.1. Fluorescence-Based GQD Sensors
2.2. Photoluminescence-Based GQD-Sensors
2.3. Chemiluminescence-Based GQD-Sensors
2.4. Electrochemiluminescence-Based GQD-Sensors
2.5. Fluorescence Resonance Energy Transfer (FRET)-Based GQD Sensors
3. Electrochemical GQD Sensors in Biomedical Diagnostics
- Implantation of the desired biomolecule.
- Blockage of the un-reacted sites.
- Determination of the target biomolecule.
3.1. Voltammetric GQD-Sensors
3.2. Amperometric GQD Sensors
3.3. Impedimetric GQD Sensors
4. Photoelectrochemical and Miscellaneous GQD Sensors in Biomedical Diagnostics
5. Summary, Critical Issues and Future Directions
Author Contributions
Funding
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stewart, J.; Manmathan, G.; Wilkinson, P. Primary prevention of cardiovascular disease: A review of contemporary guidance and literature. JRSM Cardiovasc. Dis. 2017, 6, 204800401668721. [Google Scholar] [CrossRef] [Green Version]
- Ho, K.J. Cardiovascular diseases. Nutr. Asp. Aging 2018, 2, 75–100. [Google Scholar]
- Clark, L.C.; Lyons, C. Electrode Systems for Continuous Monitoring in Cardiovascular Surgery. Ann. N. Y. Acad. Sci. 1962, 102, 29–45. [Google Scholar] [CrossRef] [PubMed]
- Sharma, T.K.; Ramanathan, R.; Rakwal, R.; Agrawal, G.K.; Bansal, V. Moving forward in plant food safety and security through NanoBioSensors: Adopt or adapt biomedical technologies? Proteomics 2015, 1680–1692. [Google Scholar] [CrossRef] [PubMed]
- Saylan, Y.; Erdem, Ö.; Ünal, S.; Denizli, A. An alternative medical diagnosis method: Biosensors for virus detection. Biosensors 2019, 9, 65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pohanka, M. Current Trends in the Biosensors for Biological Warfare Agents Assay. Materials 2019, 12, 2303. [Google Scholar] [CrossRef] [Green Version]
- Lv, M.; Liu, Y.; Geng, J.; Kou, X.; Xin, Z.; Yang, D. Engineering nanomaterials-based biosensors for food safety detection. Biosens. Bioelectron. 2018, 106, 122–128. [Google Scholar] [CrossRef]
- Hernandez-Vargas, G.; Sosa-Hernández, J.E.; Saldarriaga-Hernandez, S.; Villalba-Rodríguez, A.M.; Parra-Saldivar, R.; Iqbal, H.M.N. Electrochemical biosensors: A solution to pollution detection with reference to environmental contaminants. Biosensors 2018, 8, 29. [Google Scholar] [CrossRef] [Green Version]
- Griffin, S. Biosensors for Cancer Detection Applications. Missouri S&T’s Peer to Peer 2017, 1, 6. [Google Scholar]
- Altintas, Z.; Fakanya, W.M.; Tothill, I.E. Cardiovascular disease detection using bio-sensing techniques. Talanta 2014, 128, 177–186. [Google Scholar] [CrossRef] [PubMed]
- Jurado-Sánchez, B. Nanoscale biosensors based on self-propelled objects. Biosensors 2018, 8, 59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Altintas, Z.; Akgun, M.; Kokturk, G.; Uludag, Y. A fully automated microfluidic-based electrochemical sensor for real-time bacteria detection. Biosens. Bioelectron. 2018, 100, 541–548. [Google Scholar] [CrossRef] [PubMed]
- Puiu, M.; Bala, C. Peptide-based biosensors: From self-assembled interfaces to molecular probes in electrochemical assays. Bioelectrochemistry 2018, 120, 66–75. [Google Scholar] [CrossRef]
- Rocchitta, G.; Spanu, A.; Babudieri, S.; Latte, G.; Madeddu, G.; Galleri, G.; Nuvoli, S.; Bagella, P.; Demartis, M.I.; Fiore, V.; et al. Enzyme biosensors for biomedical applications: Strategies for safeguarding analytical performances in biological fluids. Sensors 2016, 16, 780. [Google Scholar] [CrossRef] [Green Version]
- Gupta, S.; Smith, T.; Banaszak, A.; Boeckl, J. Graphene quantum dots electrochemistry and sensitive electrocatalytic glucose sensor development. Nanomaterials 2017, 7, 301. [Google Scholar] [CrossRef]
- Grabowska, I.; Sharma, N.; Vasilescu, A.; Iancu, M.; Badea, G.; Boukherroub, R.; Ogale, S.; Szunerits, S. Electrochemical Aptamer-Based Biosensors for the Detection of Cardiac Biomarkers. ACS Omega 2018, 3, 12010–12018. [Google Scholar] [CrossRef] [Green Version]
- Savas, S.; Ersoy, A.; Gulmez, Y.; Kilic, S.; Levent, B.; Altintas, Z. Nanoparticle enhanced antibody and DNA biosensors for sensitive detection of Salmonella. Materials 2018, 11, 1541. [Google Scholar] [CrossRef] [Green Version]
- Waffo, A.F.T.; Yesildag, C.; Caserta, G.; Katz, S.; Zebger, I.; Lensen, M.C.; Wollenberger, U.; Scheller, F.W.; Altintas, Z. Fully electrochemical MIP sensor for artemisinin. Sens. Actuators B Chem. 2018, 275, 163–173. [Google Scholar] [CrossRef]
- Abdin, M.J.; Altintas, Z.; Tothill, I.E. In silico designed nanoMIP based optical sensor for endotoxins monitoring. Biosens. Bioelectron. 2015, 67, 177–183. [Google Scholar] [CrossRef]
- Mollarasouli, F.; Kurbanoglu, S.; Ozkan, S.A. The Role of Electrochemical Immunosensors in Clinical Analysis. Biosensors 2019, 9, 86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gharatape, A.; Khosroushahi, A.Y. Optical Biomarker-based Biosensors for Cancer/Infectious. Appl. Immunohistochem. Mol. Morphol. 2019, 27, 278–286. [Google Scholar] [CrossRef] [PubMed]
- Pohanka, M. Overview of piezoelectric biosensors, immunosensors and DNA sensors and their applications. Materials 2018, 11, 448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sayed, M.; Gul, M.; Shah, N.S.; Khan, J.A.; Khan, Z.U.H.; Rehman, F.; Khan, A.R.; Rauf, S.; Arandiyan, H.; Yang, C.P. In-situ dual applications of ionic liquid coated Co2+ and Fe3+ co-doped TiO2: Superior photocatalytic degradation of ofloxacin at pilot scale level and enhanced peroxidase like activity for calorimetric biosensing. J. Mol. Liq. 2019, 282, 275–285. [Google Scholar] [CrossRef]
- Boverhof, D.R.; Bramante, C.M.; Butala, J.H.; Clancy, S.F.; Lafranconi, M.; West, J.; Gordon, S.C. Comparative assessment of nanomaterial de fi nitions and safety evaluation considerations. Regul. Toxicol. Pharmacol. 2015, 73, 137–150. [Google Scholar]
- Nasrollahzadeh, M.; Sajadi, S.M.; Sajjadi, M.; Issaabadi, Z. Applications of Nanotechnology in Daily Life. Interface Sci. Technol. 2019, 28, 113–143. [Google Scholar]
- Lombardo, D.; Kiselev, M.A.; Caccamo, M.T. Smart Nanoparticles for Drug Delivery Application: Development of Versatile Nanocarrier Platforms in Biotechnology and Nanomedicine. J. Nanomater. 2019, 2019. [Google Scholar] [CrossRef]
- Gupta, R.; Xie, H. Nanoparticles in Daily Life: Applications, Toxicity and Regulations. J. Environ. Pathol. Toxicol. Oncol. 2018, 37, 209–230. [Google Scholar] [CrossRef]
- Pirzada, M.; Altintas, Z. Nanomaterials for Healthcare Biosensing Applications. Sensors 2019, 19, 5311. [Google Scholar] [CrossRef] [Green Version]
- Baer, D.R.; Engelhard, M.H.; Johnson, G.E.; Laskin, J.; Lai, J.; Mueller, K.; Thevuthasan, S.; Wang, H.; Washton, N.; Elder, A.; et al. challenging opportunities Surface characterization of nanomaterials and nanoparticles: Important needs and challenging opportunities. J. Vac. Sci. Technol. A 2014, 050820, 1–34. [Google Scholar]
- Rodríguez-López, J.L.; Montejano-Carrizales, J.M.; Palomares-Báez, J.P.; Barrón-Escobar, H.; Velázquez-Salazar, J.J.; Cabrera-Trujillo, J.M.; José-Yacamán, M. Size Effect and Shape Stability of Nanoparticles. Key Eng. Mater. 2010, 444, 47–68. [Google Scholar] [CrossRef]
- Khan, I.; Saeed, K.; Khan, I. Nanoparticles: Properties, applications and toxicities. Arab. J. Chem. 2017. [Google Scholar] [CrossRef]
- Wernsdorfer, W.; Orozco, E.B.; Barbara, B.; Benoit, A.; Mailly, D.; Demoncy, N.; Pascard, H.; Polytechnique, E.; Nakano, H.; Corporation, T.; et al. Magnetization Reversal in Individual1 Nanoparticles Macroscopic Quantum Tunneling of Magnetization. IEEE Trans. Magn. 1998, 34, 973–978. [Google Scholar] [CrossRef]
- Tonelli, D.; Scavetta, E.; Gualandi, I. Electrochemical Deposition of Nanomaterials for Electrochemical Sensing. Sensors 2019, 19, 1186. [Google Scholar] [CrossRef] [Green Version]
- Piperno, A.; Scala, A.; Mazzaglia, A.; Neri, G.; Pennisi, R.; Sciortino, M.T.; Grassi, G. Cellular signaling pathways activated by functional graphene nanomaterials. Int. J. Mol. Sci. 2018, 19, 3365. [Google Scholar] [CrossRef] [Green Version]
- Kokkinos, C. Electrochemical DNA Biosensors Based on Labeling with Nanoparticles. Nanomaterials 2019, 9, 1361. [Google Scholar] [CrossRef] [Green Version]
- Campuzano, S.; Paloma, Y.; Pingarr, M. Carbon Dots and Graphene Quantum Dots in Electrochemical Biosensing. Nanomaterials 2019, 9, 634. [Google Scholar] [CrossRef] [Green Version]
- Alizadeh Zeinabad, H.; Ghourchian, H.; Falahati, M.; Fathipour, M.; Azizi, M.; Boutorabi, S.M. Ultrasensitive interdigitated capacitance immunosensor using gold nanoparticles. Nanotechnology 2018, 29, 26. [Google Scholar] [CrossRef]
- Idris, A.O.; Mabuba, N.; Arotiba, O.A. An alpha-fetoprotein electrochemical immunosensor based on a carbon/gold bi-nanoparticle platform. Anal. Methods 2018, 10, 5649–5658. [Google Scholar] [CrossRef]
- Bohli, N.; Belkilani, M.; Mora, L.; Abdelghani, A. Antibody-functionalised gold nanoparticles-based impedimetric immunosensor: Detection methods for better sensitivity. Micro Nano Lett. 2019, 14, 629–633. [Google Scholar] [CrossRef]
- Wang, X.; Gao, D.; Li, M.; Li, H.; Li, C.; Wu, X.; Yang, B. CVD graphene as an electrochemical sensing platform for simultaneous detection of biomolecules. Sci. Rep. 2017, 7, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Raj, A.; John, S.A. Graphene-Modified Electrochemical Sensors. Graphene-Based Electrochem. Sens. Biomol. 2019, 1–41. [Google Scholar] [CrossRef]
- Bahadir, E.B.; Sezgintürk, M.K. Applications of graphene in electrochemical sensing and biosensing. TrAC Trends Anal. Chem. 2016, 76, 1–14. [Google Scholar] [CrossRef]
- Nikolaev, K.G.; Ermolenko, Y.E.; Offenhäusser, A.; Ermakov, S.S.; Mourzina, Y.G. Multisensor systems by electrochemical nanowire assembly for the analysis of aqueous solutions. Front. Chem. 2018, 6, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oliveira, T.M.B.F.; Morais, S. New generation of electrochemical sensors based on multi-walled carbon nanotubes. Appl. Sci. 2018, 8, 1925. [Google Scholar] [CrossRef] [Green Version]
- Bettazzi, F.; Natale, A.R.; Torres, E.; Palchetti, I. Glyphosate determination by coupling an immuno-magnetic assay with electrochemical sensors. Sensors 2018, 18, 2965. [Google Scholar] [CrossRef] [Green Version]
- Helali, S.; Martelet, C.; Abdelghani, A.; Maaref, M.A.; Jaffrezic-Renault, N. A disposable immunomagnetic electrochemical sensor based on functionalised magnetic beads on gold surface for the detection of atrazine. Electrochim. Acta 2006, 51, 5182–5186. [Google Scholar] [CrossRef]
- Wang, Y.; Myers, M.; Staser, J.A. Electrochemical UV sensor using carbon quantum dot/graphene semiconductor. J. Electrochem. Soc. 2018, 165, H3001–H3007. [Google Scholar] [CrossRef] [Green Version]
- Faridbod, F.; Sanati, A.L. Graphene Quantum Dots in Electrochemical Sensors/Biosensors. Curr. Anal. Chem. 2018, 15, 103–123. [Google Scholar] [CrossRef]
- Nanomaterials definition matters. Nat. Nanotechnol. 2019, 14, 193. Available online: https://0-www-nature-com.brum.beds.ac.uk/articles/s41565-019-0412-3 (accessed on 5 March 2019). [CrossRef] [Green Version]
- Kurniawan, F.; Al Kiswiyah, N.S.; Madurani, K.A.; Tominaga, M. Electrochemical sensor based on single-walled carbon nanotubes-modified gold electrode for uric acid detection. J. Electrochem. Soc. 2018, 165, B515–B522. [Google Scholar] [CrossRef]
- Liang, J.; Zheng, Y.; Liu, Z. Nanowire-based Cu electrode as electrochemical sensor for detection of nitrate in water. Sens. Actuators B Chem. 2016, 232, 336–344. [Google Scholar] [CrossRef]
- Sun, H.; Wu, L.; Wei, W.; Qu, X. Recent advances in graphene quantum dots for sensing. Mater. Today 2013, 16, 433–442. [Google Scholar] [CrossRef]
- Pedrero, M.; Campuzano, S.; Pingarrón, J.M. Quantum dots as components of electrochemical sensing platforms for the detection of environmental and food pollutants: A review. J. AOAC Int. 2017, 100, 950–961. [Google Scholar] [CrossRef] [PubMed]
- Mistry, K.K.; Layek, K.; Mahapatra, A.; RoyChaudhuri, C.; Sahab, H. A review on amperometric-type immunosensors based on screen-printed electrodes. Analyst 2014, 139, 2289–2311. [Google Scholar] [CrossRef]
- Zeng, Z.; Xiao, F.X.; Phan, H.; Chen, S.; Yu, Z.; Wang, R.; Nguyen, T.Q.; Yang Tan, T.T. Unraveling the cooperative synergy of zero-dimensional graphene quantum dots and metal nanocrystals enabled by layer-by-layer assembly. J. Mater. Chem. A 2018, 6, 1700–1713. [Google Scholar] [CrossRef]
- Hasanzadeh, M.; Shadjou, N. What are the reasons for low use of graphene quantum dots in immunosensing of cancer biomarkers? Mater. Sci. Eng. C 2017, 71, 1313–1326. [Google Scholar] [CrossRef]
- Fan, Z.; Li, S.; Yuan, F.; Fan, L. Fluorescent Graphene Quantum Dots for Biosensing and Bioimaging. RSC Adv. 2015, 5, 19773–19789. [Google Scholar] [CrossRef]
- Xie, R.; Wang, Z.; Zhou, W.; Liu, Y.; Fan, L.; Li, Y.; Li, X. Graphene quantum dots as smart probes for biosensing. Anal. Methods 2016, 8, 4001–4006. [Google Scholar] [CrossRef]
- Tachi, S.; Morita, H.; Takahashi, M.; Okabayashi, Y.; Hosokai, T.; Sugai, T.; Kuwahara, S. Quantum Yield Enhancement in Graphene Quantum Dots via Esterification with Benzyl Alcohol. Sci. Rep. 2019, 9, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Roushani, M.; Valipour, A. The potentiality of graphene quantum dots functionalized by nitrogen and thiol-doped (GQDs-N-S) to stabilize the antibodies in designing of human chorionic gonadotropin immunosensor. Nanochem. Res. 2019, 4, 20–26. [Google Scholar]
- Ma, F.; Li, C.C.; Zhang, C. yang Development of quantum dot-based biosensors: Principles and applications. J. Mater. Chem. B 2018, 6, 6173–6190. [Google Scholar] [CrossRef]
- Chen, W.; Lv, G.; Hu, W.; Li, D.; Chen, S.; Dai, Z. Synthesis and applications of graphene quantum dots: A review. Nanotechnol. Rev. 2018, 7, 157–185. [Google Scholar] [CrossRef]
- Rakovich, A.; Rakovich, T. Semiconductor: Versus graphene quantum dots as fluorescent probes for cancer diagnosis and therapy applications. J. Mater. Chem. B 2018, 6, 2690–2712. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.; Chen, J.F.; Dai, L. Recent advances in graphene quantum dots for fluorescence bioimaging from cells through tissues to animals. Part. Part. Syst. Charact. 2015, 32, 515–523. [Google Scholar] [CrossRef]
- Choi, S.H. Unique properties of graphene quantum dots and their applications in photonic/electronic devices. J. Phys. D Appl. Phys. 2017, 50, 1–10. [Google Scholar] [CrossRef]
- Savas, S.; Altintas, Z. Graphene quantum dots as nanozymes for electrochemical sensing of yersinia enterocolitica in milk and human serum. Materials 2019, 12, 2189. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Rui, M.; Song, J.; Shen, Z.; Zeng, H. Carbon and Graphene Quantum Dots for Optoelectronic and Energy Devices: A Review. Adv. Funct. Mater. 2015, 25, 4929–4947. [Google Scholar] [CrossRef]
- Zheng, P.; Wu, N. Fluorescence and Sensing Applications of Graphene Oxide and Graphene Quantum Dots: A Review. Chem. An Asian J. 2017, 12, 2343–2353. [Google Scholar] [CrossRef]
- Wang, L.; Zhu, S.; Wang, H.; Qu, S.; Zhang, Y.; Zhang, J.; Chen, Q.; Al, W.E.T. Common Origin of Green Luminescence in Carbon Nanodots and Graphene Quantum Dots. Am. Chem. Soc. 2014, 8, 2541–2547. [Google Scholar] [CrossRef]
- Shen, J.; Zhu, Y.; Yang, X.; Li, C. ChemComm Graphene quantum dots: Emergent nanolights for bioimaging, sensors, catalysis and photovoltaic devices. R. Soc. Chem. 2012, 46, 3686–3699. [Google Scholar]
- Yan, X.; Cui, X.; Li, L.S. Synthesis of large, stable colloidal graphene quantum dots with tunable size. J. Am. Chem. Soc. 2010, 132, 5944–5945. [Google Scholar] [CrossRef] [PubMed]
- Tian, P.; Tang, L.; Teng, K.S.; Lau, S.P. Graphene quantum dots from chemistry to applications. Mater. Today Chem. 2018, 10, 221–258. [Google Scholar] [CrossRef]
- Bacon, M.; Bradley, S.J.; Nann, T. Graphene quantum dots. Part. Part. Syst. Charact. 2014, 31, 415–428. [Google Scholar] [CrossRef]
- Karimzadeh, A.; Hasanzadeh, M.; Shadjou, N.; Guardia, M. de la Optical bio(sensing) using nitrogen doped graphene quantum dots: Recent advances and future challenges. TrAC Trends Anal. Chem. 2018, 108, 110–121. [Google Scholar] [CrossRef]
- Gupta, S.; Kaushal, A.; Kumar, A.; Kumar, D. Ultrasensitive transglutaminase based nanosensor for early detection of celiac disease in human. Int. J. Biol. Macromol. 2017, 105, 905–911. [Google Scholar] [CrossRef]
- Tuteja, S.K.; Chen, R.; Kukkar, M.; Song, C.K.; Mutreja, R.; Singh, S.; Paul, A.K.; Lee, H.; Kim, K.H.; Deep, A.; et al. A label-free electrochemical immunosensor for the detection of cardiac marker using graphene quantum dots (GQDs). Biosens. Bioelectron. 2016, 86, 548–556. [Google Scholar] [CrossRef]
- Yang, Y.; Liu, Q.; Liu, Y.; Cui, J.; Liu, H.; Wang, P.; Li, Y.; Chen, L.; Zhao, Z.; Dong, Y. A novel label-free electrochemical immunosensor based on functionalized nitrogen-doped graphene quantum dots for carcinoembryonic antigen detection. Biosens. Bioelectron. 2017, 90, 31–38. [Google Scholar] [CrossRef]
- Mansuriya, B.D.; Altintas, Z. Graphene Quantum Dot-Based Electrochemical Immunosensors for Biomedical Applications. Materials 2019, 13, 96. [Google Scholar] [CrossRef] [Green Version]
- Chikkaveeraiah, B.V.; Bhirde, A.A.; Morgan, N.Y.; Eden, H.S.; Chen, X. Electrochemical immunosensors for detection of cancer protein biomarkers. ACS Nano 2012, 6, 6546–6561. [Google Scholar] [CrossRef] [Green Version]
- Serafín, V.; Valverde, A.; Garranzo-Asensio, M.; Barderas, R.; Campuzano, S.; Yáñez-Sedeño, P.; Pingarrón, J.M. Simultaneous amperometric immunosensing of the metastasis-related biomarkers IL-13Rα2 and CDH-17 by using grafted screen-printed electrodes and a composite prepared from quantum dots and carbon nanotubes for signal amplification. Microchim. Acta 2019, 186. [Google Scholar] [CrossRef] [PubMed]
- Damborský, P.; Švitel, J.; Katrlík, J. Optical biosensors. Essays Biochem. 2016, 60, 91–100. [Google Scholar] [PubMed] [Green Version]
- Peltomaa, R.; Glahn-Martínez, B.; Benito-Peña, E.; Moreno-Bondi, M.C. Optical Biosensors for Label-Free Detection of Small Molecules. Sensors 2018, 18, 4216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Altintas, Z.; Uludag, Y.; Gurbuz, Y.; Tothill, I. Development of surface chemistry for surface plasmon resonance based sensors for the detection of proteins and DNA molecules. Anal. Chim. Acta 2012, 712, 138–144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pawula, M.; Altintas, Z.; Tothill, I.E. SPR detection of cardiac troponin T for acute myocardial infarction. Talanta 2016, 146, 823–830. [Google Scholar] [CrossRef] [PubMed]
- Zanchetta, G.; Lanfranco, R.; Giavazzi, F.; Bellini, T.; Buscaglia, M. Emerging applications of label-free optical biosensors. Nanophotonics 2017, 6, 627–645. [Google Scholar] [CrossRef]
- Huertas, C.S.; Calvo-Lozano, O.; Mitchell, A.; Lechuga, L.M. Advanced Evanescent-Wave Optical Biosensors for the Detection of Nucleic Acids: An Analytic Perspective. Front. Chem. 2019, 7, 1–25. [Google Scholar] [CrossRef] [Green Version]
- Nawrot, W.; Drzozga, K.; Baluta, S.; Cabaj, J.; Malecha, K. A fluorescent biosensors for detection vital body fluids’ agents. Sensors 2018, 18, 2357. [Google Scholar] [CrossRef] [Green Version]
- Strianese, M.; Maria, S.; Giuseppe, R.; Labella, T.; Claudio, P.; D’Auria, S. Spectroscopic Methods of Analysis; Bujalowski, W.M., Ed.; Methods in Molecular Biology; Springer: New York, NY, USA, 2012; Volume 875, ISBN 978-1-61779-805-4. [Google Scholar]
- Ligler, F.S. Fluorescence-Based Optical Biosensors. In Biophotonics; Springer: Berlin/Heidelberg, Germany, 2008; Volume 1, pp. 199–215. [Google Scholar]
- Girigoswami, K.; Akhtar, N. Nanobiosensors and fluorescence based biosensors: An overview. Int. J. Nano Dimens. 2019, 10, 1–17. [Google Scholar]
- Li, C.; Zhang, J.; Xiong, Q.; Lorenzini, G.; Yue, Y. The pH Effect on Thermal Response of Fluorescence Spectroscopy of Graphene Quantum Dots for Nanoscale Thermal Characterization. J. Eng. Thermophys. 2018, 27, 345–356. [Google Scholar] [CrossRef]
- Li, N.; Li, R.; Li, Z.; Yang, Y.; Wang, G.; Gu, Z. Pentaethylenehexamine and histidine-functionalized graphene quantum dots for ultrasensitive fluorescence detection of microRNA with target and molecular beacon double cycle amplification strategy. Sens. Actuators B Chem. 2019, 283, 666–676. [Google Scholar] [CrossRef]
- Laurenti, M.; Paez-Perez, M.; Algarra, M.; Alonso-Cristobal, P.; Lopez-Cabarcos, E.; Mendez-Gonzalez, D.; Rubio-Retama, J. Enhancement of the Upconversion Emission by Visible-to-Near-Infrared Fluorescent Graphene Quantum Dots for miRNA Detection. ACS Appl. Mater. Interfaces 2016, 8, 12644–12651. [Google Scholar] [CrossRef] [PubMed]
- Kermani, H.A.; Hosseini, M.; Dadmehr, M.; Hosseinkhani, S.; Ganjali, M.R. DNA methyltransferase activity detection based on graphene quantum dots using fluorescence and fluorescence anisotropy. Sens. Actuators B Chem. 2017, 241, 217–223. [Google Scholar] [CrossRef]
- Wang, G.L.; Fang, X.; Wu, X.M.; Hu, X.L.; Li, Z.J. Label-free and ratiometric detection of nuclei acids based on graphene quantum dots utilizing cascade amplification by nicking endonuclease and catalytic G-quadruplex DNAzyme. Biosens. Bioelectron. 2016, 81, 214–220. [Google Scholar] [CrossRef]
- Sun, L.; Li, S.; Ding, W.; Yao, Y.; Yang, X.; Yao, C. Fluorescence detection of cholesterol using a nitrogen-doped graphene quantum dot/chromium picolinate complex-based sensor. J. Mater. Chem. B 2017, 5, 9006–9014. [Google Scholar] [CrossRef]
- Ryu, J.; Lee, E.; Lee, K.; Jang, J. A graphene quantum dots based fluorescent sensor for anthrax biomarker detection and its size dependence. J. Mater. Chem. B 2015, 3, 4865–4870. [Google Scholar] [CrossRef]
- Feng, L.L.; Wu, Y.X.; Zhang, D.L.; Hu, X.X.; Zhang, J.; Wang, P.; Song, Z.L.; Zhang, X.B.; Tan, W. Near Infrared Graphene Quantum Dots-Based Two-Photon Nanoprobe for Direct Bioimaging of Endogenous Ascorbic Acid in Living Cells. Anal. Chem. 2017, 89, 4077–4084. [Google Scholar] [CrossRef]
- Liu, H.; Na, W.; Liu, Z.; Chen, X.; Su, X. A novel turn-on fluorescent strategy for sensing ascorbic acid using graphene quantum dots as fluorescent probe. Biosens. Bioelectron. 2017, 92, 229–233. [Google Scholar] [CrossRef]
- Na, W.; Li, N.; Xingguang, S. Enzymatic growth of single-layer MnO2 nanosheets in situ: Application to detect alkaline phosphatase and ascorbic acid in the presence of sulfanilic acid functionalized graphene quantum dots. Sens. Actuators B Chem. 2018, 274, 172–179. [Google Scholar] [CrossRef]
- Liu, J.J.; Tang, D.; Chen, Z.; Yan, X.; Zhong, Z.; Kang, L.; Yao, J. Chemical redox modulated fluorescence of nitrogen-doped graphene quantum dots for probing the activity of alkaline phosphatase. Biosens. Bioelectron. 2017, 94, 271–277. [Google Scholar] [CrossRef]
- Zhu, Y.; Wang, G.; Jiang, H.; Chen, L.; Zhang, X. One-step ultrasonic synthesis of graphene quantum dots with high quantum yield and their application in sensing alkaline phosphatase. Chem. Commun. 2015, 51, 948–951. [Google Scholar] [CrossRef] [PubMed]
- Cui, F.; Ji, J.; Sun, J.; Wang, J.; Wang, H.; Zhang, Y.; Ding, H.; Lu, Y.; Xu, D.; Sun, X. A novel magnetic fluorescent biosensor based on graphene quantum dots for rapid, efficient, and sensitive separation and detection of circulating tumor cells. Anal. Bioanal. Chem. 2019, 411, 985–995. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Gao, X.; Song, F.; Wang, C.; Chu, F.; Wu, S. A sensing approach for dopamine determination by boronic acid-functionalized molecularly imprinted graphene quantum dots composite. Appl. Surf. Sci. 2017, 423, 810–816. [Google Scholar] [CrossRef]
- Weng, S.; Liang, D.; Qiu, H.; Liu, Z.; Lin, Z.; Zheng, Z.; Liu, A.; Chen, W.; Lin, X. A unique turn-off fluorescent strategy for sensing dopamine based on formed polydopamine (pDA) using graphene quantum dots (GQDs) as fluorescent probe. Sens. Actuators B Chem. 2015, 221, 7–14. [Google Scholar] [CrossRef] [Green Version]
- Zhou, X.; Wang, A.; Yu, C.; Wu, S.; Shen, J. Facile Synthesis of Molecularly Imprinted Graphene Quantum Dots for the Determination of Dopamine with Affinity-Adjustable. ACS Appl. Mater. Interfaces 2015, 7, 11741–11747. [Google Scholar] [CrossRef]
- Zhou, X.; Ma, P.; Wang, A.; Yu, C.; Qian, T.; Wu, S.; Shen, J. Dopamine fluorescent sensors based on polypyrrole/graphene quantum dots core/shell hybrids. Biosens. Bioelectron. 2014, 64, 404–410. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Zhao, L.; Lan, C.; Zhao, S. Graphene quantum dots as effective probes for label-free fluorescence detection of dopamine. Sens. Actuators B Chem. 2016, 223, 246–251. [Google Scholar] [CrossRef]
- Tashkhourian, J.; Dehbozorgi, A. Determination of dopamine in the presence of ascorbic and uric acids by fluorometric method using graphene quantum dots. Spectrosc. Lett. 2016, 49, 319–325. [Google Scholar] [CrossRef]
- Xiaoyan, Z.; Yuanyuan, J.; Zaijun, L.; Zhiguo, G.; Guangli, W. Improved activity and thermo-stability of the horse radish peroxidase with graphene quantum dots and its application in fluorometric detection of hydrogen peroxide. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2016, 165, 106–113. [Google Scholar] [CrossRef]
- Zhao, W.; Li, Y.; Yang, S.; Chen, Y.; Zheng, J.; Liu, C.; Qing, Z.; Li, J.; Yang, R. Target-Activated Modulation of Dual-Color and Two-Photon Fluorescence of Graphene Quantum Dots for in Vivo Imaging of Hydrogen Peroxide. Anal. Chem. 2016, 88, 4833–4840. [Google Scholar] [CrossRef]
- Qu, Z.; Na, W.; Nie, Y.; Su, X. A novel fluorimetric sensing strategy for highly sensitive detection of phytic acid and hydrogen peroxide. Anal. Chim. Acta 2018, 1039, 74–81. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Shen, J.; Huang, Y.; Liu, Z.; Zhuang, H. Graphene quantum dots and enzyme-coupled biosensor for highly sensitive determination of hydrogen peroxide and glucose. Int. J. Mol. Sci. 2018, 19, 1696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, Y.; Wang, X.; Sun, J.; Jiao, S.; Chen, H.; Gao, F.; Wang, L. Fluorescent blood glucose monitor by hemin-functionalized graphene quantum dots based sensing system. Anal. Chim. Acta 2014, 810, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Gu, Z.; Lei, W.; Wang, W.; Xia, X.; Hao, Q. Graphene quantum dots as a fluorescent sensing platform for highly efficient detection of copper(II) ions. Sens. Actuators B Chem. 2014, 190, 516–522. [Google Scholar] [CrossRef]
- Qian, Z.S.; Shan, X.Y.; Chai, L.J.; Chen, J.R.; Feng, H. A fluorescent nanosensor based on graphene quantum dots-aptamer probe and graphene oxide platform for detection of lead (II) ion. Biosens. Bioelectron. 2015, 68, 225–231. [Google Scholar] [CrossRef] [PubMed]
- Niu, X.; Zhong, Y.; Chen, R.; Wang, F.; Liu, Y.; Luo, D. A “turn-on” fluorescence sensor for Pb2+ detection based on graphene quantum dots and gold nanoparticles. Sens. Actuators B Chem. 2018, 255, 1577–1581. [Google Scholar] [CrossRef]
- Anh, N.T.N.; Chowdhury, A.D.; Doong, R.A. Highly sensitive and selective detection of mercury ions using N, S-codoped graphene quantum dots and its paper strip based sensing application in wastewater. Sens. Actuators B Chem. 2017, 252, 1169–1178. [Google Scholar] [CrossRef]
- Xiaoyan, Z.; Zhangyi, L.; Zaijun, L. Fabrication of valine-functionalized graphene quantum dots and its use as a novel optical probe for sensitive and selective detection of Hg2+. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2017, 171, 415–424. [Google Scholar] [CrossRef]
- Bian, S.; Shen, C.; Qian, Y.; Liu, J.; Xi, F.; Dong, X. Facile synthesis of sulfur-doped graphene quantum dots as fluorescent sensing probes for Ag+ ions detection. Sens. Actuators B Chem. 2017, 242, 231–237. [Google Scholar] [CrossRef]
- Zhao, X.E.; Lei, C.; Gao, Y.; Gao, H.; Zhu, S.; Yang, X.; You, J.; Wang, H. A ratiometric fluorescent nanosensor for the detection of silver ions using graphene quantum dots. Sens. Actuators B Chem. 2017, 253, 239–246. [Google Scholar] [CrossRef]
- Ran, X.; Sun, H.; Pu, F.; Ren, J.; Qu, X. Ag Nanoparticle-decorated graphene quantum dots for label-free, rapid and sensitive detection of Ag+ and biothiols. Chem. Commun. 2013, 49, 1079–1081. [Google Scholar] [CrossRef]
- He, L.; Yang, L.; Zhu, H.; Dong, W.; Ding, Y.; Zhu, J.J. A highly sensitive biosensing platform based on upconversion nanoparticles and graphene quantum dots for the detection of Ag+. Methods Appl. Fluoresc. 2017, 5, 1–11. [Google Scholar] [CrossRef]
- Ju, J.; Chen, W. Synthesis of highly fluorescent nitrogen-doped graphene quantum dots for sensitive, label-free detection of Fe (III) in aqueous media. Biosens. Bioelectron. 2014, 58, 219–225. [Google Scholar] [CrossRef]
- Guo, R.; Zhou, S.; Li, Y.; Li, X.; Fan, L.; Voelcker, N.H. Rhodamine-Functionalized Graphene Quantum Dots for Detection of Fe3+ in Cancer Stem Cells. ACS Appl. Mater. Interfaces 2015, 7, 23958–23966. [Google Scholar] [CrossRef] [PubMed]
- Van Tam, T.; Trung, N.B.; Kim, H.R.; Chung, J.S.; Choi, W.M. One-pot synthesis of N-doped graphene quantum dots as a fluorescent sensing platform for Fe3+ ions detection. Sens. Actuators B Chem. 2014, 202, 568–573. [Google Scholar] [CrossRef]
- Ge, S.; He, J.; Ma, C.; Liu, J.; Xi, F.; Dong, X. One-step synthesis of boron-doped graphene quantum dots for fluorescent sensors and biosensor. Talanta 2019, 199, 581–589. [Google Scholar] [CrossRef] [PubMed]
- Dutta Chowdhury, A.; Doong, R.A. Highly Sensitive and Selective Detection of Nanomolar Ferric Ions Using Dopamine Functionalized Graphene Quantum Dots. ACS Appl. Mater. Interfaces 2016, 8, 21002–21010. [Google Scholar] [CrossRef]
- Li, S.; Li, Y.; Cao, J.; Zhu, J.; Fan, L.; Li, X. Sulfur-doped graphene quantum dots as a novel fluorescent probe for highly selective and sensitive detection of Fe3+. Anal. Chem. 2014, 86, 10201–10207. [Google Scholar] [CrossRef]
- Wu, Z.; Li, W.; Chen, J.; Yu, C. A graphene quantum dot-based method for the highly sensitive and selective fluorescence turn on detection of biothiols. Talanta 2014, 119, 538–543. [Google Scholar] [CrossRef]
- Zhang, R.; Sun, J.; Ji, J.; Pi, F.; Xiao, Y.; Zhang, Y.; Sun, X. A novel “OFF-ON” biosensor based on nanosurface energy transfer between gold nanocrosses and graphene quantum dots for intracellular ATP sensing and tracking. Sens. Actuators B Chem. 2019, 282, 910–916. [Google Scholar] [CrossRef]
- Ganganboina, A.B.; Dutta Chowdhury, A.; Doong, R.A. N-Doped Graphene Quantum Dots-Decorated V2O5 Nanosheet for Fluorescence Turn Off-On Detection of Cysteine. ACS Appl. Mater. Interfaces 2018, 10, 614–624. [Google Scholar] [CrossRef]
- Yan, Y.J.; He, X.W.; Li, W.Y.; Zhang, Y.K. Nitrogen-doped graphene quantum dots-labeled epitope imprinted polymer with double templates via the metal chelation for specific recognition of cytochrome c. Biosens. Bioelectron. 2017, 91, 253–261. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Sun, J.; He, P.; Deng, X.; Wang, Z.; Hu, C.; Ding, G.; Xie, X. Selenium doped graphene quantum dots as an ultrasensitive redox fluorescent switch. Chem. Mater. 2015, 27, 2004–2011. [Google Scholar] [CrossRef]
- Li, X.; Zhu, S.; Xu, B.; Ma, K.; Zhang, J.; Yang, B.; Tian, W. Self-assembled graphene quantum dots induced by cytochrome c: A novel biosensor for trypsin with remarkable fluorescence enhancement. Nanoscale 2013, 5, 7776–7779. [Google Scholar] [CrossRef] [PubMed]
- Qu, Z.; Na, W.; Liu, X.; Liu, H.; Su, X. A novel fluorescence biosensor for sensitivity detection of tyrosinase and acid phosphatase based on nitrogen-doped graphene quantum dots. Anal. Chim. Acta 2018, 997, 52–59. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Than, A.; Wang, X.; Xu, S.; Sun, L.; Duan, H.; Xu, C.; Chen, P. Ultrasensitive Profiling of Metabolites Using Tyramine-Functionalized Graphene Quantum Dots. ACS Nano 2016, 10, 3622–3629. [Google Scholar] [CrossRef]
- Benítez-Martínez, S.; Caballero-Díaz, E.; Valcárcel, M. Development of a biosensing system for tacrine based on nitrogen-doped graphene quantum dots and acetylcholinesterase. Analyst 2016, 141, 2688–2695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, F.; Zeng, Q.; Lai, Z.; Cheng, Z.; Ruan, G. Silicon doped graphene quantum dots combined with ruthenium(III) ions as a fluorescent probe for turn-on detection of triclosan. New J. Chem. 2019, 43, 12907–12915. [Google Scholar] [CrossRef]
- Zou, F.; Zhou, H.; Van Tan, T.; Kim, J.; Koh, K.; Lee, J. Dual-Mode SERS-Fluorescence Immunoassay Using Graphene Quantum Dot Labeling on One-Dimensional Aligned Magnetoplasmonic Nanoparticles. ACS Appl. Mater. Interfaces 2015, 7, 12168–12175. [Google Scholar] [CrossRef]
- Kaur, M.; Mehta, S.K.; Kansal, S.K. A fluorescent probe based on nitrogen doped graphene quantum dots for turn off sensing of explosive and detrimental water pollutant, TNP in aqueous medium. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2017, 180, 37–43. [Google Scholar] [CrossRef]
- Cai, N.; Tan, L.; Li, Y.; Xia, T.; Hu, T.; Su, X. Biosensing platform for the detection of uric acid based on graphene quantum dots and G-quadruplex/hemin DNAzyme. Anal. Chim. Acta 2017, 965, 96–102. [Google Scholar] [CrossRef]
- Liu, Y.; Cao, N.; Gui, W.; Ma, Q. Nitrogen-doped graphene quantum dots-based fluorescence molecularly imprinted sensor for thiacloprid detection. Talanta 2018, 183, 339–344. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Huang, H.; Ma, Y.; Tong, J. Highly sensitive fluorescent detection of dihydroxybenzene based on graphene quantum dots. Sens. Actuators B Chem. 2014, 205, 227–233. [Google Scholar] [CrossRef]
- Shi, J.; Lyu, J.; Tian, F.; Yang, M. A fluorescence turn-on biosensor based on graphene quantum dots (GQDs) and molybdenum disulfide (MoS2) nanosheets for epithelial cell adhesion molecule (EpCAM) detection. Biosens. Bioelectron. 2017, 93, 182–188. [Google Scholar] [CrossRef] [PubMed]
- Khataee, A.; Hassanzadeh, J.; Kohan, E. Specific quantification of atropine using molecularly imprinted polymer on graphene quantum dots. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2018, 205, 614–621. [Google Scholar] [CrossRef] [PubMed]
- Jiang, D.; Ni, D.; Liu, F.; Zhang, L.; Liu, L.; Pu, X. A fluorescent imaging assay of cast in renal disease based on graphene quantum dots and Fe3O4 nanoparticles. Clin. Chim. Acta 2016, 454, 94–101. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Na, W.; Liu, Q.; Su, X. A novel label-free fluorescent sensor for highly sensitive detection of bleomycin based on nitrogen-doped graphene quantum dots. Anal. Chim. Acta 2018, 1028, 45–49. [Google Scholar] [CrossRef]
- Shao, T.; Zhang, P.; Tang, L.; Zhuo, S.; Zhu, C. Highly sensitive enzymatic determination of urea based on the pH-dependence of the fluorescence of graphene quantum dots. Microchim. Acta 2015, 182, 1431–1437. [Google Scholar] [CrossRef]
- Zhou, T.; Halder, A.; Sun, Y. Fluorescent nanosensor based on molecularly imprinted polymers coated on graphene quantum dots for fast detection of antibiotics. Biosensors 2018, 8, 82. [Google Scholar] [CrossRef] [Green Version]
- Huang, S.; Wang, L.; Huang, C.; Su, W.; Xiao, Q. Label-free and ratiometric fluorescent nanosensor based on amino-functionalized graphene quantum dots coupling catalytic G-quadruplex/hemin DNAzyme for ultrasensitive recognition of human telomere DNA. Sens. Actuators B Chem. 2017, 245, 648–655. [Google Scholar] [CrossRef]
- Zhou, Y.; Qu, Z.B.; Zeng, Y.; Zhou, T.; Shi, G. A novel composite of graphene quantum dots and molecularly imprinted polymer for fluorescent detection of paranitrophenol. Biosens. Bioelectron. 2014, 52, 317–323. [Google Scholar] [CrossRef]
- Zhu, S.; Yan, X.; Sun, J.; Zhao, X.E.; Wang, X. A novel and sensitive fluorescent assay for artemisinin with graphene quantum dots based on inner filter effect. Talanta 2019, 200, 163–168. [Google Scholar] [CrossRef] [PubMed]
- Mehrzad-Samarin, M.; Faridbod, F.; Ganjali, M.R. A luminescence nanosensor for Ornidazole detection using graphene quantum dots entrapped in silica molecular imprinted polymer. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2019, 206, 430–436. [Google Scholar] [CrossRef] [PubMed]
- Sahub, C.; Tuntulani, T.; Nhujak, T.; Tomapatanaget, B. Effective biosensor based on graphene quantum dots via enzymatic reaction for directly photoluminescence detection of organophosphate pesticide. Sens. Actuators B Chem. 2018, 258, 88–97. [Google Scholar] [CrossRef]
- Zor, E.; Morales-Narváez, E.; Zamora-Gálvez, A.; Bingol, H.; Ersoz, M.; Merkoçi, A. Graphene quantum dots-based photoluminescent sensor: A multifunctional composite for pesticide detection. ACS Appl. Mater. Interfaces 2015, 7, 20272–20279. [Google Scholar] [CrossRef]
- Zhao, H.; Chang, Y.; Liu, M.; Gao, S.; Yu, H.; Quan, X. A universal immunosensing strategy based on regulation of the interaction between graphene and graphene quantum dots. Chem. Commun. 2013, 49, 234–236. [Google Scholar] [CrossRef]
- Mondal, T.K.; Dinda, D.; Saha, S.K. Nitrogen, sulphur co-doped graphene quantum dot: An excellent sensor for nitroexplosives. Sens. Actuators B Chem. 2018, 257, 586–593. [Google Scholar] [CrossRef]
- He, Y.; Sun, J.; Feng, D.; Chen, H.; Gao, F.; Wang, L. Graphene quantum dots: Highly active bifunctional nanoprobes for nonenzymatic photoluminescence detection of hydroquinone. Biosens. Bioelectron. 2015, 74, 418–422. [Google Scholar] [CrossRef]
- Zhang, Q.; Song, C.; Zhao, T.; Fu, H.W.; Wang, H.Z.; Wang, Y.J.; Kong, D.M. Photoluminescent sensing for acidic amino acids based on the disruption of graphene quantum dots/europium ions aggregates. Biosens. Bioelectron. 2015, 65, 204–210. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, L.; Liang, R.P.; Bai, J.M.; Qiu, J.D. Using graphene quantum dots as photoluminescent probes for protein kinase sensing. Anal. Chem. 2013, 85, 9148–9155. [Google Scholar] [CrossRef]
- Ju, J.; Zhang, R.; He, S.; Chen, W. Nitrogen-doped graphene quantum dots-based fluorescent probe for the sensitive turn-on detection of glutathione and its cellular imaging. RSC Adv. 2014, 4, 52583–52589. [Google Scholar] [CrossRef]
- Safardoust-Hojaghan, H.; Salavati-Niasari, M.; Amiri, O.; Hassanpour, M. Preparation of highly luminescent nitrogen doped graphene quantum dots and their application as a probe for detection of Staphylococcus aureus and E. coli. J. Mol. Liq. 2017, 241, 1114–1119. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, Z.Y.; Liang, R.P.; Li, Y.H.; Qiu, J.D. Boron-doped graphene quantum dots for selective glucose sensing based on the ‘abnormal’ aggregation-induced photoluminescence enhancement. Anal. Chem. 2014, 86, 4423–4430. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Peng, D.; Liang, R.P.; Qiu, J.D. Graphene Quantum Dots Assembled with Metalloporphyrins for ‘turn on’ Sensing of Hydrogen Peroxide and Glucose. Chem. A Eur. J. 2015, 21, 9343–9348. [Google Scholar] [CrossRef] [PubMed]
- Shehab, M.; Ebrahim, S.; Soliman, M. Graphene quantum dots prepared from glucose as optical sensor for glucose. J. Lumin. 2017, 184, 110–116. [Google Scholar] [CrossRef]
- Huang, H.; Liao, L.; Xu, X.; Zou, M.; Liu, F.; Li, N. The electron-transfer based interaction between transition metal ions and photoluminescent graphene quantum dots (GQDs): A platform for metal ion sensing. Talanta 2013, 117, 152–157. [Google Scholar] [CrossRef]
- Ananthanarayanan, A.; Wang, X.; Routh, P.; Sana, B.; Lim, S.; Kim, D.H.; Lim, K.H.; Li, J.; Chen, P. Facile synthesis of graphene quantum dots from 3D graphene and their application for Fe3+ sensing. Adv. Funct. Mater. 2014, 24, 3021–3026. [Google Scholar] [CrossRef]
- Zhou, L.; Geng, J.; Liu, B. Graphene quantum dots from polycyclic aromatic hydrocarbon for bioimaging and sensing of Fe3+ and hydrogen peroxide. Part. Part. Syst. Charact. 2013, 30, 1086–1092. [Google Scholar] [CrossRef]
- Wang, L.; Li, W.; Wu, B.; Li, Z.; Wang, S.; Liu, Y.; Pan, D.; Wu, M. Facile synthesis of fluorescent graphene quantum dots from coffee grounds for bioimaging and sensing. Chem. Eng. J. 2016, 300, 75–82. [Google Scholar] [CrossRef]
- Sun, H.; Gao, N.; Wu, L.; Ren, J.; Wei, W.; Qu, X. Highly photoluminescent amino-functionalized graphene quantum dots used for sensing copper ions. Chem. A Eur. J. 2013, 19, 13362–13368. [Google Scholar] [CrossRef]
- Bai, J.M.; Zhang, L.; Liang, R.P.; Qiu, J.D. Graphene quantum dots combined with europium ions as photoluminescent probes for phosphate sensing. Chem. A Eur. J. 2013, 19, 3822–3826. [Google Scholar] [CrossRef]
- Patra, S.; Roy, E.; Choudhary, R.; Tiwari, A.; Madhuri, R.; Sharma, P.K. Graphene quantum dots decorated CdS doped graphene oxide sheets in dual action mode: As initiator and platform for designing of nimesulide imprinted polymer. Biosens. Bioelectron. 2017, 89, 627–635. [Google Scholar] [CrossRef]
- Aboul-Enein, H.Y.; Stefan, R.I.; Van Staden, J.F. Chemiluminescence-based (bio)sensors—An overview. Crit. Rev. Anal. Chem. 1999, 29, 323–331. [Google Scholar] [CrossRef]
- Baeyens, W.R.G.; Schulman, S.G.; Calokerinos, A.C.; Zhao, Y.; García Campaña, A.M.; Nakashima, K.; De Keukeleire, D. Chemiluminescence-based detection: Principles and analytical applications in flowing streams and in immunoassays. J. Pharm. Biomed. Anal. 1998, 17, 941–953. [Google Scholar] [CrossRef]
- Amjadi, M.; Manzoori, J.L.; Hallaj, T. Chemiluminescence of graphene quantum dots and its application to the determination of uric acid. J. Lumin. 2014, 153, 73–78. [Google Scholar] [CrossRef]
- Amjadi, M.; Jalili, R. Molecularly imprinted polymer-capped nitrogen-doped graphene quantum dots as a novel chemiluminescence sensor for selective and sensitive determination of doxorubicin. RSC Adv. 2016, 6, 86736–86743. [Google Scholar] [CrossRef]
- Chen, H.; Wang, Q.; Shen, Q.; Liu, X.; Li, W.; Nie, Z.; Yao, S. Nitrogen doped graphene quantum dots based long-persistent chemiluminescence system for ascorbic acid imaging. Biosens. Bioelectron. 2017, 91, 878–884. [Google Scholar] [CrossRef]
- Hassanzadeh, J.; Khataee, A. Ultrasensitive chemiluminescent biosensor for the detection of cholesterol based on synergetic peroxidase-like activity of MoS2 and graphene quantum dots. Talanta 2018, 178, 992–1000. [Google Scholar] [CrossRef]
- Al-Ogaidi, I.; Gou, H.; Aguilar, Z.P.; Guo, S.; Melconian, A.K.; Al-Kazaz, A.K.A.; Meng, F.; Wu, N. Detection of the ovarian cancer biomarker CA-125 using chemiluminescence resonance energy transfer to graphene quantum dots. Chem. Commun. 2014, 50, 1344–1346. [Google Scholar] [CrossRef]
- Hamtak, M.; Hosseini, M.; Fotouhi, L.; Aghazadeh, M. A new electrochemiluminescence biosensor for the detection of glucose based on polypyrrole/polyluminol/Ni(OH)2-C3N4/glucose oxidase-modified graphite electrode. Anal. Methods 2018, 10, 5723–5730. [Google Scholar] [CrossRef]
- Zhuo, Y.; Wang, H.J.; Lei, Y.M.; Zhang, P.; Liu, J.L.; Chai, Y.Q.; Yuan, R. Electrochemiluminescence biosensing based on different modes of switching signals. Analyst 2018, 143, 3230–3248. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, X.; Xu, Z.; Chai, Y.; Wang, H.; Yuan, R. An ultrasensitive electrochemiluminescence biosensor for multiple detection of microRNAs based on a novel dual circuit catalyzed hairpin assembly. Chem. Commun. 2018, 54, 10148–10151. [Google Scholar] [CrossRef] [PubMed]
- Gross, E.M.; Maddipati, S.S.; Snyder, S.M. A review of electrogenerated chemiluminescent biosensors for assays in biological matrices. Bioanalysis 2016, 8, 2071–2089. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nie, Y.; Yuan, X.; Zhang, P.; Chai, Y.Q.; Yuan, R. Versatile and Ultrasensitive Electrochemiluminescence Biosensor for Biomarker Detection Based on Nonenzymatic Amplification and Aptamer-Triggered Emitter Release. Anal. Chem. 2019, 91, 3452–3458. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Zhou, T.; Huang, R. Recent advances in Electrochemiluminescence sensors for pathogenic bacteria detection. Micromachines 2019, 10, 532. [Google Scholar] [CrossRef] [Green Version]
- Lin, Y.; Huang, X.; Zhang, Y.; Chen, D.; Wang, J.; Luo, F.; Guo, L.; Qiu, B.; Lin, Z. Electrochemiluminescence Biosensor for the Detection of the Folate Receptor in HeLa Cells Based on Hyperbranched Rolling Circle Amplification and Terminal Protection. ChemElectroChem 2019, 6, 827–833. [Google Scholar] [CrossRef]
- Lu, J.; Yan, M.; Ge, L.; Ge, S.; Wang, S.; Yan, J. Biosensors and Bioelectronics Electrochemiluminescence of blue-luminescent graphene quantum dots and its application in ultrasensitive aptasensor for adenosine triphosphate detection. Biosens. Bioelectron. 2013, 47, 271–277. [Google Scholar] [CrossRef]
- Wu, D.; Liu, Y.; Wang, Y.; Hu, L.; Ma, H.; Wang, G.; Wei, Q. Label-free Electrochemiluminescent Immunosensor for Detection of Prostate Specific Antigen based on Aminated Graphene Quantum Dots and Carboxyl Graphene Quantum Dots. Sci. Rep. 2016, 6, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.; Liu, W.; Ma, C.; Zhang, Y.; Wang, X.; Yu, J.; Song, X. Gold-silver nanocomposite-functionalized graphene based electrochemiluminescence immunosensor using graphene quantum dots coated porous PtPd nanochains as labels. Electrochim. Acta 2014, 123, 470–476. [Google Scholar] [CrossRef]
- Kmezic, S.; Radenkovic, D.; Pejovic, I.; Antic, A.; Bajec, Đ. The significance of tumor markers, CA 19-9 and CEA, in the pancreatic cancer staging and evaluation of surgical resectability. Int. Hepato-Pancreato-Biliary Assoc. 2016, 18, e372. [Google Scholar] [CrossRef] [Green Version]
- Grunnet, M.; Sorensen, J.B. Lung Cancer Carcinoembryonic antigen (CEA) as tumor marker in lung cancer. Lung Cancer 2012, 76, 138–143. [Google Scholar] [CrossRef]
- Cetean, S.; Laszlo, I.; Constantin, A.; Căinap, S. Classic tumor markers in gastric cancer. Current standards and limitations. Clujul Med. 2015, 88, 111–115. [Google Scholar]
- Saito, G.; Sadahiro, S.; Kamata, H.; Miyakita, H. Monitoring of Serum Carcinoembryonic Antigen Levels after Curative Resection of Colon Cancer: Cutoff Values Determined according to Preoperative Levels Enhance the Diagnostic. Oncology 2017, 92, 276–282. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Lee, S. The Roles of Carcinoembryonic Antigen in Liver Metastasis and Therapeutic Approaches. Gastroenterol. Res. Pract. 2017, 2017, 1–11. [Google Scholar] [CrossRef]
- Cho, W.K.; Choi, D.H.; Park, H.C.; Park, W.; Yu, J.I.; Cho, B.; Yun, S.H.; Lee, W.Y. Elevated CEA is associated with worse survival in recurrent rectal cancer. Oncotarget 2017, 8, 105936–105941. [Google Scholar] [CrossRef] [Green Version]
- Press, D. Diagnostic and prognostic value of carcinoembryonic antigen in pancreatic cancer: A systematic review and meta-analysis. OncoTargets Ther. 2017, 10, 4591–4598. [Google Scholar]
- Asad-ur-rahman, F.N.U.; Saif, M.W. Elevated Level of Serum Carcinoembryonic Antigen (CEA) and Search for a Malignancy: A Case Report. Cureus 2016, 8, 8–11. [Google Scholar] [CrossRef] [Green Version]
- Altintas, Z.; Tothill, I. Biomarkers and biosensors for the early diagnosis of lung cancer. Sens. Actuators B Chem. 2013, 188, 988–998. [Google Scholar] [CrossRef]
- Nie, G.; Wang, Y.; Tang, Y.; Zhao, D.; Guo, Q. A graphene quantum dots based electrochemiluminescence immunosensor for carcinoembryonic antigen detection using poly(5-formylindole)/reduced graphene oxide nanocomposite. Biosens. Bioelectron. 2018, 101, 123–128. [Google Scholar] [CrossRef]
- Chen, S.; Chen, X.; Zhang, L.; Gao, J.; Ma, Q. Electrochemiluminescence Detection of Escherichia coli O157:H7 Based on a Novel Polydopamine Surface Imprinted Polymer Biosensor. ACS Appl. Mater. Interfaces 2017, 9, 5430–5436. [Google Scholar] [CrossRef]
- Lu, Q.; Wei, W.; Zhou, Z.; Zhou, Z.; Liu, S. Electrochemiluminescence resonance energy transfer between graphene quantum dots and gold nanoparticles for DNA damage detection †. Analyst 2014, 2404–2410. [Google Scholar] [CrossRef]
- Zhou, C.; Chen, Y.; You, X.; Dong, Y.; Chi, Y. An Electrochemiluminescent Biosensor Based on Interactions between a Graphene Quantum Dot À Sulfite Co-reactant System and Hydrogen Peroxide. Chem. Electrochem. 2017, 4, 1783–1789. [Google Scholar] [CrossRef] [Green Version]
- Tian, K.; Nie, F.; Luo, K.; Zheng, X.; Zheng, J. A sensitive electrochemiluminescence glucose biosensor based on graphene quantum dot prepared from graphene oxide sheets and hydrogen peroxide. J. Electroanal. Chem. 2017, 2017, 1–30. [Google Scholar] [CrossRef]
- Liang, R.; Qiu, W.; Zhao, H.; Xiang, C.; Qiu, J. Electrochemiluminescence Resonance Energy Transfer Between Graphene Quantum Dots and Graphene Oxide for Sensitive Protein Kinase Activity and Inhibitor Sensing. Anal. Chim. Acta 2016, 2016, 1–25. [Google Scholar] [CrossRef]
- Chen, S.; Chen, X.; Xia, T.; Ma, Q. Biosensors and Bioelectronics A novel electrochemiluminescence sensor for the detection of nitroaniline based on the nitrogen-doped graphene quantum dots. Biosens. Bioelectron. 2016, 85, 903–908. [Google Scholar] [CrossRef]
- Yan, Y.; Liu, Q.; Dong, X.; Hao, N.; Chen, S.; You, T.; Mao, H.; Wang, K. Copper (I) oxide nanospheres decorated with graphene quantum dots display improved electrocatalytic activity for enhanced luminol electrochemiluminescence. Microchim. Acta 2016, 183, 1591–1599. [Google Scholar] [CrossRef]
- Liu, Q.; Wang, K.; Huan, J.; Zhu, G.; Qian, J. Graphene quantum dots enhanced electrochemiluminescence of cadmium sul fi de nanocrystals for ultrasensitive determination of pentachlorophenol. Analyst 2014, 139, 2912–2918. [Google Scholar] [CrossRef]
- Du, X.; Jiang, D.; Liu, Q.; Zhu, G.; Wang, K. Fabrication of graphene oxide decorated with nitrogen-doped graphene quantum dots and its enhanced electrochemiluminescence for ultrasensitive detection of pentachlorophenol. Analyst 2015, 140, 1253–1259. [Google Scholar] [CrossRef]
- Dong, Y.; Tian, W.; Ren, S.; Dai, R.; Chi, Y.; Chen, G. Graphene Quantum Dots/L-Cysteine Coreactant Electrochemiluminescence System and Its Application in Sensing Lead(II) Ions Yongqiang. ACS Appl. Mater. Interfaces 2014, 6, 1646–1651. [Google Scholar] [CrossRef]
- Jie, G.; Zhou, Q.; Jie, G. Talanta Graphene quantum dots-based electrochemiluminescence detection of DNA using multiple cycling amplification strategy. Talanta 2019, 194, 658–663. [Google Scholar] [CrossRef]
- Chen, H.; Li, W.; Wang, Q.; Jin, X.; Nie, Z.; Yao, S. Electrochimica Acta Nitrogen doped graphene quantum dots based single-luminophor generated dual-potential electrochemiluminescence system for ratiometric sensing of Co2+ ion. Electrochim. Acta 2016, 214, 94–102. [Google Scholar] [CrossRef]
- Wang, X.; Liu, L.; Wang, Z.; Dai, Z. Highly sensitive electrochemiluminescent DNA biosensor based on hydrazide-modified graphene quantum dots and hemin/G-quadruplex DNAzyme. J. Electroanal. Chem. 2016, 781, 351–355. [Google Scholar] [CrossRef]
- Dong, S.; Bi, Q.; Qiao, C.; Sun, Y.; Zhang, X.; Lu, X.; Zhao, L. Electrochemical sensor for discrimination tyrosine enantiomers using graphene quantum dots and β-cyclodextrins composites. Talanta 2017, 173, 94–100. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Qian, J.; Wang, K.; Hua, M.; Liu, Q.; Hao, N.; You, T.; Huang, X. Nitrogen-Doped Graphene Quantum Dots@SiO2 Nanoparticles as Electrochemiluminescence and Fluorescence Signal Indicators for Magnetically Controlled Aptasensor with Dual Detection Channels. ACS Appl. Mater. Interfaces 2015, 7, 26865–26873. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Fang, G.; Wang, X.; Zhang, F.; Liu, J.; Zheng, W.; Wang, S. Electrochemiluminescent graphene quantum dots enhanced by MoS2 as sensing platform: A novel molecularly imprinted electrochemiluminescence sensor for 2-methyl-4-chlorophenoxyacetic acid assay. Electrochim. Acta 2017, 228, 107–113. [Google Scholar] [CrossRef]
- Lou, J.; Liu, S.; Tu, W.; Dai, Z. Graphene quantums dots combined with endonuclease cleavage and bidentate chelation for highly sensitive electrochemiluminescent DNA biosensing. Anal. Chem. 2015, 87, 1145–1151. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Zhuo, Y.; Chang, Y.; Yuan, R.; Chai, Y. Electrochemiluminescent Graphene Quantum Dots as a Sensing Platform: A Dual Amplification for MicroRNA Assay. Anal. Chem. 2015, 87, 10385–10391. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Wu, H.; Shang, P.; Zeng, X.; Chi, Y. Immobilizing water-soluble graphene quantum dots with gold nanoparticles for a low potential electrochemiluminescence immunosensor. Nanoscale 2015, 7, 16366–16371. [Google Scholar] [CrossRef]
- Zhang, T.; Zhao, H.; Fan, G.; Li, Y.; Li, L.; Quan, X. Electrolytic exfoliation synthesis of boron doped graphene quantum dots: A new luminescent material for electrochemiluminescence detection of oncogene microRNA-20a. Electrochim. Acta 2016, 190, 1150–1158. [Google Scholar] [CrossRef]
- Li, J.J.; Shang, L.; Jia, L.P.; Ma, R.N.; Zhang, W.; Jia, W.L.; Wang, H.S.; Xu, K.H. An ultrasensitive electrochemiluminescence sensor for the detection of HULC based on Au@Ag/GQDs as a signal indicator. J. Electroanal. Chem. 2018, 824, 114–120. [Google Scholar] [CrossRef]
- Zadran, S.; Standley, S.; Wong, K.; Otiniano, E.; Amighi, A.; Baudry, M. Fluorescence resonance energy transfer (FRET)-based biosensors: Visualizing cellular dynamics and bioenergetics. Appl. Microbiol. Biotechnol. 2012, 96, 895–902. [Google Scholar] [CrossRef]
- Qian, Z.S.; Shan, X.Y.; Chai, L.J.; Ma, J.J.; Chen, J.R.; Feng, H. DNA nanosensor based on biocompatible graphene quantum dots and carbon nanotubes. Biosens. Bioelectron. 2014, 60, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Qian, Z.S.; Shan, X.Y.; Chai, L.J.; Ma, J.J.; Chen, J.R.; Feng, H. A universal fluorescence sensing strategy based on biocompatible graphene quantum dots and graphene oxide for the detection of DNA. Nanoscale 2014, 6, 5671–5674. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.L. Hydrolysis of Phosphate Esters Catalyzed by Inorganic Iron Oxide Nanoparticles Acting as Biocatalysts. Astrobiology 2018, 18, 294–310. [Google Scholar] [CrossRef] [PubMed]
- Kirschenbaum, A.; Izadmehr, S.; Yao, S.; O’Connor-Chapman, K.L.; Huang, A.; Gregoriades, E.M.; Yakar, S.; Levine, A.C. Prostatic acid phosphatase alters the RANKL/OPG system and induces osteoblastic prostate cancer bone metastases. Endocrinology 2016, 157, 4526–4533. [Google Scholar] [CrossRef] [Green Version]
- Meana, C.; García-Rostán, G.; Peña, L.; Lordén, G.; Cubero, Á.; Orduña, A.; Győrffy, B.; Balsinde, J.; Balboa, M.A. The phosphatidic acid phosphatase lipin-1 facilitates inflammation-driven colon carcinogenesis. JCI insight 2018, 3, 1–18. [Google Scholar] [CrossRef]
- Na, W.; Liu, Q.; Sui, B.; Hu, T.; Su, X. Highly sensitive detection of acid phosphatase by using a graphene quantum dots-based förster resonance energy transfer. Talanta 2016, 161, 469–475. [Google Scholar] [CrossRef]
- Li, Y.; Sun, L.; Qian, J.; Wang, C.; Liu, Q.; Han, E.; Hao, N.; Zhang, L.; Cai, J.; Wang, K. A homogeneous assay for highly sensitive detection of CaMV35S promoter in transgenic soybean by förster resonance energy transfer between nitrogen-doped graphene quantum dots and Ag nanoparticles. Anal. Chim. Acta 2016, 948, 90–97. [Google Scholar] [CrossRef]
- Kong, L.; Li, Y.; Ma, C.; Liu, B.; Tan, L. Sensitive immunoassay of von Willebrand factor based on fluorescence resonance energy transfer between graphene quantum dots and Ag@Au nanoparticles. Colloids Surf. B Biointerfaces 2018, 165, 286–292. [Google Scholar] [CrossRef]
- Poon, C.Y.; Li, Q.; Zhang, J.; Li, Z.; Dong, C.; Lee, A.W.M.; Chan, W.H.; Li, H.W. FRET-based modified graphene quantum dots for direct trypsin quantification in urine. Anal. Chim. Acta 2016, 917, 64–70. [Google Scholar] [CrossRef]
- Yan, X.; Song, Y.; Zhu, C.; Song, J.; Du, D.; Su, X.; Lin, Y. Graphene Quantum Dot-MnO2 Nanosheet Based Optical Sensing Platform: A Sensitive Fluorescence ‘turn Off-On’ Nanosensor for Glutathione Detection and Intracellular Imaging. ACS Appl. Mater. Interfaces 2016, 8, 21990–21996. [Google Scholar] [CrossRef]
- Yan, X.; Zhao, X.E.; Sun, J.; Zhu, S.; Lei, C.; Li, R.; Gong, P.; Ling, B.; Wang, R.; Wang, H. Probing glutathione reductase activity with graphene quantum dots and gold nanoparticles system. Sens. Actuators B Chem. 2018, 263, 27–35. [Google Scholar] [CrossRef]
- Sun, J.; Cui, F.; Zhang, R.; Gao, Z.; Ji, J.; Ren, Y.; Pi, F.; Zhang, Y.; Sun, X. Comet-like Heterodimers ‘gold Nanoflower @Graphene Quantum Dots’ Probe with FRET ‘off’ to DNA Circuit Signal ‘on’ for Sensing and Imaging MicroRNA in Vitro and in Vivo. Anal. Chem. 2018, 90, 11538–11547. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, Y.; Zhao, D.; Zeng, D.; Xia, J.; Aldalbahi, A.; Wang, C.; San, L.; Fan, C.; Zuo, X.; et al. Universal Fluorescence Biosensor Platform Based on Graphene Quantum Dots and Pyrene-Functionalized Molecular Beacons for Detection of MicroRNAs. ACS Appl. Mater. Interfaces 2015, 7, 16152–16156. [Google Scholar] [CrossRef]
- Fan, L.; Hu, Y.; Wang, X.; Zhang, L.; Li, F.; Han, D.; Li, Z.; Zhang, Q.; Wang, Z.; Niu, L. Fluorescence resonance energy transfer quenching at the surface of graphene quantum dots for ultrasensitive detection of TNT. Talanta 2012, 101, 192–197. [Google Scholar] [CrossRef]
- Shi, J.; Chan, C.; Pang, Y.; Ye, W.; Tian, F.; Lyu, J.; Zhang, Y.; Yang, M. A fluorescence resonance energy transfer (FRET) biosensor based on graphene quantum dots (GQDs) and gold nanoparticles (AuNPs) for the detection of mecA gene sequence of Staphylococcus aureus. Biosens. Bioelectron. 2015, 67, 595–600. [Google Scholar] [CrossRef]
- Bhatnagar, D.; Kumar, V.; Kumar, A.; Kaur, I. Graphene quantum dots FRET based sensor for early detection of heart attack in human. Biosens. Bioelectron. 2016, 79, 495–499. [Google Scholar] [CrossRef]
- Stradiotto, N.R.; Yamanaka, H.; Zanoni, M.V.B. Electrochemical sensors: A powerful tool in analytical chemistry. J. Braz. Chem. Soc. 2003, 14, 159–173. [Google Scholar] [CrossRef] [Green Version]
- Lim, S.A.; Ahmed, M.U. Electrochemical immunosensors and their recent nanomaterial-based signal amplification strategies: A review. RCS Adv. 2016. [Google Scholar] [CrossRef]
- Kokkinos, C.; Economou, A.; Prodromidis, M.I. Electrochemical immunosensors: Critical survey of different architectures and transduction strategies. Trends Anal. Chem. 2015. [Google Scholar] [CrossRef]
- Rama, E.C.; Costa-García, A. Screen-printed Electrochemical Immunosensors for the Detection of Cancer and Cardiovascular Biomarkers. Electroanalysis 2016, 28, 1700–1715. [Google Scholar] [CrossRef]
- Cho, I.H.; Lee, J.; Kim, J.; Kang, M.S.; Paik, J.K.; Ku, S.; Cho, H.M.; Irudayaraj, J.; Kim, D.H. Current technologies of electrochemical immunosensors: Perspective on signal amplification. Sensors 2018, 18, 207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Faridbod, F.; Gupta, V.K.; Zamani, H.A. Electrochemical Sensors and Biosensors. Int. J. Electrochem. 2011, 24, 717. [Google Scholar] [CrossRef] [Green Version]
- Dhahi, T.H.S.; Bin Hashim, U.D.A.; Ahmed, N.M.; Mat Taib, A. A review on the electrochemical sensors and biosensors composed of nanogaps as sensing material. J. Optoelectron. Adv. Mater. 2010, 12, 1857–1862. [Google Scholar]
- Khristunova, Y.; Korotkova, E.; Kratochvil, B.; Barek, J.; Dorozhko, E.; Vyskocil, V.; Plotnikov, E.; Voronova, O.; Sidelnikov, V. Preparation and Investigation of Silver Nanoparticle—Antibody Bioconjugates for. Sensors 2019, 19, 2103. [Google Scholar] [CrossRef] [Green Version]
- Thevenot, D.R.; Toth, K.; Durst, R.A.; Wilson, G.S. Electrochemical biosensors: Recommended defnitions and classification. Pure Appl. Chem. 1999, 71, 2333–2348. [Google Scholar] [CrossRef] [Green Version]
- Mehrotra, P. Biosensors and their applications—A review. J. Oral Biol. Craniofacial Res. 2016, 6, 153–159. [Google Scholar] [CrossRef] [Green Version]
- Yan, Q.; Yang, Y.; Tan, Z.; Liu, Q.; Liu, H.; Wang, P.; Chen, L.; Zhang, D.; Li, Y.; Dong, Y. A label-free electrochemical immunosensor based on the novel signal amplification system of AuPdCu ternary nanoparticles functionalized polymer nanospheres. Biosens. Bioelectron. 2018, 103, 151–157. [Google Scholar] [CrossRef]
- Pan, D.; Li, G.; Hu, H.; Xue, H.; Zhang, M.; Zhu, M.; Gong, X.; Zhang, Y.; Wan, Y.; Shen, Y. Direct Immunoassay for Facile and Sensitive Detection of Small Molecule Aflatoxin B1 based on Nanobody. Chem. A Eur. J. 2018, 24, 9869–9876. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, X.; Yifeng, E.; Fang, F.; Kuang, G.; Wang, G. Sandwich Immunoassays of Multicomponent Subtrace Pathogenic DNA Based on Magnetic Fluorescent Encoded Nanoparticles. Biomed Res. Int. 2016, 2016, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Dutta, G.; Lillehoj, P.B. Wash-free, label-free immunoassay for rapid electrochemical detection of PfHRP2 in whole blood samples. Sci. Rep. 2018, 8, 1–8. [Google Scholar] [CrossRef]
- Purvis, D.; Leonardova, O.; Farmakovsky, D.; Cherkasov, V. An ultrasensitive and stable potentiometric immunosensor. Biosens. Bioelectron. 2003, 18, 1385–1390. [Google Scholar] [CrossRef]
- Farghaly, O.A.; Abdel Hameed, R.S.; Abu-Nawwas, A.A.H. Analytical application using modern electrochemical techniques. Int. J. Electrochem. Sci. 2014, 9, 3287–3318. [Google Scholar]
- Mendoza, S.; Bustos, E.; Manríquez, J.; Godínez, L.A. Voltammetric Techniques. Agric. Food Electroanal. 2015, 21–48. [Google Scholar]
- Elgrishi, N.; Rountree, K.J.; McCarthy, B.D.; Rountree, E.S.; Eisenhart, T.T.; Dempsey, J.L. A Practical Beginner’s Guide to Cyclic Voltammetry. J. Chem. Educ. 2018, 95, 197–206. [Google Scholar] [CrossRef]
- Mirceski, V.; Gulaboski, R.; Lovric, M.; Bogeski, I.; Kappl, R.; Hoth, M. Square-Wave Voltammetry: A Review on the Recent Progress. Electroanalysis 2013, 25, 2411–2422. [Google Scholar] [CrossRef] [Green Version]
- Mirceski, V.; Gulaboski, R. Recent advances in square-wave voltammetry: A review. Maced. J. Chem. Chem. Eng. 2014, 33, 1–12. [Google Scholar]
- Bard, A.J.; Faulkner, L.R. Electrochemical Methods: Fundamentals and Applications, 2nd ed.; Willey: Hoboken, NJ, USA, 2001; ISBN 9780123813749. [Google Scholar]
- Altintas, Z.; Takiden, A.; Utesch, T.; Mroginski, M.A.; Schmid, B.; Scheller, F.W.; Süssmuth, R.D. Integrated Approaches Toward High-Affinity Artificial Protein Binders Obtained via Computationally Simulated Epitopes for Protein Recognition. Adv. Funct. Mater. 2019, 29, 1–11. [Google Scholar] [CrossRef]
- Tchinda, R.; Tutsch, A.; Schmid, B.; Süssmuth, R.D.; Altintas, Z. Recognition of protein biomarkers using epitope-mediated molecularly imprinted films: Histidine or cysteine modified epitopes? Biosens. Bioelectron. 2019, 123, 260–268. [Google Scholar] [CrossRef]
- Ensafi, A.A.; Nasr-Esfahani, P.; Rezaei, B. Metronidazole determination with an extremely sensitive and selective electrochemical sensor based on graphene nanoplatelets and molecularly imprinted polymers on graphene quantum dots. Sens. Actuators B Chem. 2018, 270, 192–199. [Google Scholar] [CrossRef]
- Hatamluyi, B.; Es’haghi, Z. Electrochemical biosensing platform based on molecularly imprinted polymer reinforced by ZnO–graphene capped quantum dots for 6-mercaptopurine detection. Electrochim. Acta 2018, 283, 1170–1177. [Google Scholar] [CrossRef]
- Yola, M.L.; Atar, N. A novel detection approach for serotonin by graphene quantum dots/two-dimensional (2D) hexagonal boron nitride nanosheets with molecularly imprinted polymer. Appl. Surf. Sci. 2018, 458, 648–655. [Google Scholar] [CrossRef]
- Roushani, M.; Jalilian, Z.; Nezhadali, A. Screen printed carbon electrode sensor with thiol graphene quantum dots and gold nanoparticles for voltammetric determination of solatol. Heliyon 2019, 5, e01984. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, W.; Yang, H.; Ma, C.; Ding, Y.N.; Ge, S.; Yu, J.; Yan, M. Graphene-palladium nanowires based electrochemical sensor using ZnFe2O4-graphene quantum dots as an effective peroxidase mimic. Anal. Chim. Acta 2014, 852, 181–188. [Google Scholar] [CrossRef] [PubMed]
- Joshi, P.; Waghmode, S. Graphene quantum dot-based on-chip electrochemical DNA hybridization sensor for pancreatic cancer. Rep. Electrochem. 2016, 6, 31–40. [Google Scholar] [CrossRef] [Green Version]
- Mahmoudi-Moghaddam, H.; Tajik, S.; Beitollahi, H. A new electrochemical DNA biosensor based on modified carbon paste electrode using graphene quantum dots and ionic liquid for determination of topotecan. Microchem. J. 2019, 150, 104085. [Google Scholar] [CrossRef]
- Arvand, M.; Hemmati, S. Magnetic nanoparticles embedded with graphene quantum dots and multiwalled carbon nanotubes as a sensing platform for electrochemical detection of progesterone. Sens. Actuators B Chem. 2017, 238, 346–356. [Google Scholar] [CrossRef]
- Vasilescu, I.; Eremia, S.A.V.; Kusko, M.; Radoi, A.; Vasile, E.; Radu, G.L. Molybdenum disulphide and graphene quantum dots as electrode modifiers for laccase biosensor. Biosens. Bioelectron. 2016, 75, 232–237. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, W.; Zhang, L.; Li, J.; Su, Z.; Wei, G. Sequence-Designed Peptide Nanofibers Bridged Conjugation of Graphene Quantum Dots with Graphene Oxide for High Performance Electrochemical Hydrogen Peroxide Biosensor. Adv. Mater. Interfaces 2017, 4, 1–13. [Google Scholar] [CrossRef]
- Wang, H.; Li, R.; Li, Z. Nanohybrid of Co3O4 and histidine-functionalized graphene quantum dots for electrochemical detection of hydroquinone. Electrochim. Acta 2017, 255, 323–334. [Google Scholar] [CrossRef]
- Wang, L.; Tricard, S.; Yue, P.; Zhao, J.; Fang, J.; Shen, W. Polypyrrole and graphene quantum dots@Prussian Blue hybrid film on graphite felt electrodes: Application for amperometric determination of l-cysteine. Biosens. Bioelectron. 2016, 77, 1112–1118. [Google Scholar] [CrossRef]
- Lu, L.; Zhou, L.; Chen, J.; Yan, F.; Liu, J.; Dong, X.; Xi, F.; Chen, P. Nanochannel-Confined Graphene Quantum Dots for Ultrasensitive Electrochemical Analysis of Complex Samples. ACS Nano 2018, 12, 12673–12681. [Google Scholar] [CrossRef]
- Li, Y.; Jiang, Y.; Mo, T.; Zhou, H.; Li, Y.; Li, S. Highly selective dopamine sensor based on graphene quantum dots self-assembled monolayers modified electrode. J. Electroanal. Chem. 2016, 767, 84–90. [Google Scholar] [CrossRef]
- Arvand, M.; Abbasnejad, S.; Ghodsi, N. Graphene quantum dots decorated with Fe3O4 nanoparticles/functionalized multiwalled carbon nanotubes as a new sensing platform for electrochemical determination of l-DOPA in agricultural products. Anal. Methods 2016, 8, 5861–5868. [Google Scholar] [CrossRef]
- Hasanzadeh, M.; Hashemzadeh, N.; Shadjou, N.; Eivazi-Ziaei, J.; Khoubnasabjafari, M.; Jouyban, A. Sensing of doxorubicin hydrochloride using graphene quantum dot modified glassy carbon electrode. J. Mol. Liq. 2016, 221, 354–357. [Google Scholar] [CrossRef]
- Wang, G.; Shi, G.; Chen, X.; Yao, R.; Chen, F. A glassy carbon electrode modified with grapheme quantum dots and silver nanoparticles for simultaneous determination of guanine and adenine. Microchim. Acta 2015, 182, 315–322. [Google Scholar] [CrossRef]
- Hasanpour, F.; Nekoeinia, M.; Semnani, A.; Shojaei, S. NiMnO3 nanoparticles anchored on graphene quantum dot: Application in sensitive electroanalysis of dobutamine. Microchem. J. 2018, 142, 17–23. [Google Scholar] [CrossRef]
- Valipour, A.; Roushani, M. Using silver nanoparticle and thiol graphene quantum dots nanocomposite as a substratum to load antibody for detection of hepatitis C virus core antigen: Electrochemical oxidation of riboflavin was used as redox probe. Biosens. Bioelectron. 2017, 89, 946–951. [Google Scholar] [CrossRef]
- Cai, J.; Sun, B.; Gou, X.; Gou, Y.; Li, W.; Hu, F. A novel way for analysis of calycosin via polyaniline functionalized graphene quantum dots fabricated electrochemical sensor. J. Electroanal. Chem. 2018, 816, 123–131. [Google Scholar] [CrossRef]
- Hasanzadeh, M.; Baghban, H.N.; Shadjou, N.; Mokhtarzadeh, A. Ultrasensitive electrochemical immunosensing of tumor suppressor protein p53 in unprocessed human plasma and cell lysates using a novel nanocomposite based on poly-cysteine/graphene quantum dots/gold nanoparticle. Int. J. Biol. Macromol. 2018, 107, 1348–1363. [Google Scholar] [CrossRef]
- Hasanzadeh, M.; Tagi, S.; Solhi, E.; Mokhtarzadeh, A.; Shadjou, N.; Eftekhari, A.; Mahboob, S. An innovative immunosensor for ultrasensitive detection of breast cancer specific carbohydrate (CA 15-3) in unprocessed human plasma and MCF-7 breast cancer cell lysates using gold nanospear electrochemically assembled onto thiolated graphene quantum dots. Int. J. Biol. Macromol. 2018, 114, 1008–1017. [Google Scholar] [CrossRef]
- Yang, M.; Javadi, A.; Gong, S. Sensitive electrochemical immunosensor for the detection of cancer biomarker using quantum dot functionalized graphene sheets as labels. Sens. Actuators B Chem. 2011, 155, 357–360. [Google Scholar] [CrossRef]
- Mollarasouli, F.; Serafín, V.; Campuzano, S.; Yáñez-Sedeño, P.; Pingarrón, J.M.; Asadpour-Zeynali, K. Ultrasensitive determination of receptor tyrosine kinase with a label-free electrochemical immunosensor using graphene quantum dots-modified screen-printed electrodes. Anal. Chim. Acta 2018, 1011, 28–34. [Google Scholar] [CrossRef]
- Wang, W.; Zhao, J.; Wen, X.; Chun-Jen Lin, C.; Li, J.; Huang, Q.; Yu, Y.; Lin, S.Y.; Li, C. MicroPET/CT imaging of AXL downregulation by HSP90 inhibition in triple-negative breast cancer. Contrast Media Mol. Imaging 2017, 2017. [Google Scholar] [CrossRef] [Green Version]
- Tufa, L.T.; Oh, S.; Tran, V.T.; Kim, J.; Jeong, K.J.; Park, T.J.; Kim, H.J.; Lee, J. Electrochemical immunosensor using nanotriplex of graphene quantum dots, Fe3O4 and Ag nanoparticles for tuberculosis. Electrochim. Acta 2018, 290, 369–377. [Google Scholar] [CrossRef]
- Wang, X.; Chen, L.; Su, X.; Ai, S. Electrochemical immunosensor with graphene quantum dots and apoferritin-encapsulated Cu nanoparticles double-assisted signal amplification for detection of avian leukosis virus subgroup J. Biosens. Bioelectron. 2013, 47, 171–177. [Google Scholar] [CrossRef] [PubMed]
- Bhatnagar, D.; Kaur, I.; Kumar, A. Ultrasensitive cardiac troponin I antibody based nanohybrid sensor for rapid detection of human heart attack. Int. J. Biol. Macromol. 2017, 95, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Lakshmanakumar, M.; Nesakumar, N.; Sethuraman, S.; Rajan, K.S.; Krishnan, U.M.; Rayappan, J.B.B. Functionalized Graphene Quantum Dot Interfaced Electrochemical Detection of Cardiac Troponin I: An Antibody Free Approach. Sci. Rep. 2019, 9, 17348. [Google Scholar] [CrossRef]
- Baluta, S.; Lesiak, A.; Cabaj, J. Graphene Quantum Dots-based Electrochemical Biosensor for Catecholamine Neurotransmitters Detection. Electroanalysis 2018, 30, 1773–1782. [Google Scholar] [CrossRef]
- Ganganboina, A.B.; Doong, R. an Functionalized N-doped graphene quantum dots for electrochemical determination of cholesterol through host-guest inclusion. Microchim. Acta 2018, 185, 526. [Google Scholar] [CrossRef]
- Xiang, Q.; Huang, J.; Huang, H.; Mao, W.; Ye, Z. A label-free electrochemical platform for the highly sensitive detection of hepatitis B virus DNA using graphene quantum dots. RSC Adv. 2018, 8, 1820–1825. [Google Scholar] [CrossRef] [Green Version]
- Yola, M.L.; Atar, N. A highly efficient nanomaterial with molecular imprinting polymer: Carbon nitride nanotubes decorated with graphene quantum dots for sensitive electrochemical determination of chlorpyrifos. J. Electrochem. Soc. 2017, 164, B223–B229. [Google Scholar] [CrossRef]
- Yola, M.L.; Atar, N. Phenylethanolamine A (PEA) imprinted polymer on carbon nitride nanotubes/graphene quantum dots/core-shell nanoparticle composite for electrochemical PEA detection in Urine sample. J. Electrochem. Soc. 2018, 165, H1–H9. [Google Scholar] [CrossRef]
- Li, R.; Liu, L.; Zhu, H.; Li, Z. Synthesis of gold-palladium nanowaxberries/dodecylamine-functionalized graphene quantum dots-graphene micro-aerogel for voltammetric determination of peanut allergen Ara h 1. Anal. Chim. Acta 2018, 1008, 38–47. [Google Scholar] [CrossRef] [PubMed]
- Akyıldırım, O.; Kardaş, F.; Beytur, M.; Yüksek, H.; Atar, N.; Yola, M.L. Palladium nanoparticles functionalized graphene quantum dots with molecularly imprinted polymer for electrochemical analysis of citrinin. J. Mol. Liq. 2017, 243, 677–681. [Google Scholar] [CrossRef]
- Yu, H.W.; Zhang, Z.; Shen, T.; Jiang, J.H.; Chang, D.; Pan, H.Z. Sensitive determination of uric acid by using graphene quantum dots as a new substrate for immobilisation of uric oxidase. IET Nanobiotechnology 2018, 12, 191–195. [Google Scholar] [CrossRef]
- Bali Prasad, B.; Kumar, A.; Singh, R. Synthesis of novel monomeric graphene quantum dots and corresponding nanocomposite with molecularly imprinted polymer for electrochemical detection of an anticancerous ifosfamide drug. Biosens. Bioelectron. 2017, 94, 1–9. [Google Scholar] [CrossRef]
- Akbarnia, A.; Zare, H.R. A voltammetric assay for microRNA-25 based on the use of amino-functionalized graphene quantum dots and ss- and ds-DNAs as gene probes. Microchim. Acta 2018, 185, 503. [Google Scholar] [CrossRef]
- Rao, H.; Zhao, X.; Liu, X.; Zhong, J.; Zhang, Z.; Zou, P.; Jiang, Y.; Wang, X.; Wang, Y. A novel molecularly imprinted electrochemical sensor based on graphene quantum dots coated on hollow nickel nanospheres with high sensitivity and selectivity for the rapid determination of bisphenol S. Biosens. Bioelectron. 2018, 100, 341–347. [Google Scholar] [CrossRef]
- Mistry, K.K.; Layek, K.; Chell, T.N.; Chaudhuri, C.R.; Saha, H. Design and development of an amperometric immunosensor based on screen-printed electrodes. Anal. Methods 2016, 8, 3096–3101. [Google Scholar] [CrossRef]
- Ronkainen, N.J.; Okon, S.L. Nanomaterial-Based Electrochemical Immunosensors for Clinically Significant Biomarkers. Materials 2014, 7, 4669–4709. [Google Scholar] [CrossRef] [Green Version]
- Hu, T.; Zhang, L.; Wen, W.; Zhang, X.; Wang, S. Enzyme catalytic amplification of miRNA-155 detection with graphene quantum dot-based electrochemical biosensor. Biosens. Bioelectron. 2016, 77, 451–456. [Google Scholar] [CrossRef] [PubMed]
- Mars, A.; Hamami, M.; Bechnak, L.; Patra, D.; Raouafi, N. Curcumin-graphene quantum dots for dual mode sensing platform: Electrochemical and fluorescence detection of APOe4, responsible of Alzheimer’s disease. Anal. Chim. Acta 2018, 1036, 141–146. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Hu, T.; Bao, T.; Zhao, L.; Zeng, X.; Wen, W.; Zhang, X.; Wang, S. A label-free electrochemical biosensor for methyltransferase activity detection and inhibitor screening based on graphene quantum dot and enzyme-catalyzed reaction. J. Electroanal. Chem. 2017, 799, 327–332. [Google Scholar] [CrossRef]
- Dourandish, Z.; Beitollahi, H. Electrochemical sensing of isoproterenol using graphite screen-printed electrode modified with graphene quantum dots. Anal. Bioanal. Electrochem. 2018, 10, 192–202. [Google Scholar]
- Zhu, Y.; Lu, S.; Gowri Manohari, A.; Dong, X.; Chen, F.; Xu, W.; Shi, Z.; Xu, C. Polydopamine interconnected graphene quantum dots and gold nanoparticles for enzymeless H2O2 detection. J. Electroanal. Chem. 2017, 796, 75–81. [Google Scholar] [CrossRef]
- Ju, J.; Chen, W. In Situ Growth of Surfactant-Free Gold Nanoparticles on Nitrogen- Doped Graphene Quantum Dots for Electrochemical Detection of Hydrogen Peroxide in Biological Environments. Anal. Chem. 2015, 87, 1903–1910. [Google Scholar] [CrossRef]
- Razmi, H.; Mohammad-Rezaei, R. Graphene quantum dots as a new substrate for immobilization and direct electrochemistry of glucose oxidase: Application to sensitive glucose determination. Biosens. Bioelectron. 2013, 41, 498–504. [Google Scholar] [CrossRef]
- Mohammad-Rezaei, R.; Razmi, H. Preparation and characterization of hemoglobin immobilized on graphene quantum dots-chitosan nanocomposite as a sensitive and stable hydrogen peroxide biosensor. Sens. Lett. 2016, 14, 685–691. [Google Scholar] [CrossRef]
- Xi, J.; Xie, C.; Zhang, Y.; Wang, L.; Xiao, J.; Duan, X.; Ren, J.; Xiao, F.; Wang, S. Pd Nanoparticles Decorated N-Doped Graphene Quantum Dots@N-Doped Carbon Hollow Nanospheres with High Electrochemical Sensing Performance in Cancer Detection. ACS Appl. Mater. Interfaces 2016, 8, 22563–22573. [Google Scholar] [CrossRef]
- Muthurasu, A.; Ganesh, V. Horseradish Peroxidase Enzyme Immobilized Graphene Quantum Dots as Electrochemical Biosensors. Appl. Biochem. Biotechnol. 2014, 174, 945–959. [Google Scholar] [CrossRef]
- Jiang, Y.; Li, Y.; Li, Y.; Li, S. A sensitive enzyme-free hydrogen peroxide sensor based on a chitosan-graphene quantum dot/silver nanocube nanocomposite modified electrode. Anal. Methods 2016, 8, 2448–2455. [Google Scholar] [CrossRef]
- Roushani, M.; Abdi, Z. Novel electrochemical sensor based on graphene quantum dots/riboflavin nanocomposite for the detection of persulfate. Sens. Actuators B Chem. 2014, 201, 503–510. [Google Scholar] [CrossRef]
- Okamoto, H.; Yoshimatsu, Y.; Tomizawa, T.; Kunita, A.; Takayama, R.; Morikawa, T.; Komura, D.; Takahashi, K.; Oshima, T.; Sato, M.; et al. Interleukin-13 receptor α2 is a novel marker and potential therapeutic target for human melanoma. Sci. Rep. 2019, 9, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Serafín, V.; Valverde, A.; Martínez-García, G.; Martínez-Periñán, E.; Comba, F.; Garranzo-Asensio, M.; Barderas, R.; Yáñez-Sedeño, P.; Campuzano, S.; Pingarrón, J.M. Graphene quantum dots-functionalized multi-walled carbon nanotubes as nanocarriers in electrochemical immunosensing. Determination of IL-13 receptor A2 in colorectal cells and tumor tissues with different metastatic potential. Sens. Actuators B Chem. 2019, 711–722. [Google Scholar]
- Malekzad, H.; Hasanzadeh, M.; Shadjou, N.; Jouyban, A. Highly sensitive immunosensing of prostate specific antigen using poly cysteine caped by graphene quantum dots and gold nanoparticle: A novel signal amplification strategy. Int. J. Biol. Macromol. 2017, 105, 522–532. [Google Scholar] [CrossRef]
- Katz, E.; Willner, I. Probing Biomolecular Interactions at Conductive and Semiconductive Surfaces by Impedance Spectroscopy: Routes to Impedimetric Immunosensors, DNA-Sensors, and Enzyme Biosensors. Electroanalysis 2003, 15, 913–947. [Google Scholar]
- Bahadir, E.B.; Sezgintürk, M.K. A review on impedimetric biosensors. Artif. Cells, Nanomedicine Biotechnol. 2016, 44, 248–262. [Google Scholar] [CrossRef]
- Prodromidis, M. Impedimetric Biosensors and Immunosensors. Proc. 2nd Int. Semin. Anal. Sci. 2007, 8, 69–71. [Google Scholar]
- Daniels, J.S.; Pourmand, N. Label-free impedance biosensors: Opportunities and challenges. Electroanalysis 2007, 19, 1239–1257. [Google Scholar] [CrossRef]
- Ravalli, A.; Marrazza, G. Electrochemical-Based Biosensor Technologies in Disease Detection and Diagnostics. In Biosensors and Nanotechnology; Altintas, Z., Ed.; Wiley: Hoboken, NJ, USA, 2018; pp. 95–124. [Google Scholar]
- Masikini, M.; Mailu, S.N.; Tsegaye, A.; Njomo, N.; Molapo, K.M.; Ikpo, C.O.; Sunday, C.E.; Rassie, C.; Wilson, L.; Baker, P.G.L.; et al. A fumonisins immunosensor based on polyanilino-carbon nanotubes doped with palladium telluride quantum dots. Sensors 2015, 15, 529–546. [Google Scholar] [CrossRef]
- Bhardwaj, H.; Singh, C.; Kotnala, R.K.; Sumana, G. Graphene quantum dots-based nano-biointerface platform for food toxin detection. Anal. Bioanal. Chem. 2018, 410, 7313–7323. [Google Scholar] [CrossRef]
- Zhao, M. Direct Synthesis of Graphene Quantum Dots with Different Fluorescence Properties by Oxidation of Graphene Oxide Using Nitric Acid. Appl. Sci. 2018, 8, 1303. [Google Scholar] [CrossRef] [Green Version]
- Mehta, J.; Bhardwaj, N.; Bhardwaj, S.K.; Tuteja, S.K.; Vinayak, P.; Paul, A.K.; Kim, K.H.; Deep, A. Graphene quantum dot modified screen printed immunosensor for the determination of parathion. Anal. Biochem. 2017, 523, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Elshafey, R.; Radi, A.E. Electrochemical impedance sensor for herbicide alachlor based on imprinted polymer receptor. J. Electroanal. Chem. 2018, 813, 171–177. [Google Scholar] [CrossRef]
- Ghanbari, K.; Roushani, M.; Azadbakht, A. Ultra-sensitive aptasensor based on a GQD nanocomposite for detection of hepatitis C virus core antigen. Anal. Biochem. 2017, 534, 64–69. [Google Scholar] [CrossRef] [PubMed]
- Ghanbari, K.; Roushani, M. A novel electrochemical aptasensor for highly sensitive and quantitative detection of the streptomycin antibiotic. Bioelectrochemistry 2018, 120, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Ganganboina, A.B.; Doong, R. Graphene Quantum Dots Decorated Gold-Polyaniline Nanowire for Impedimetric Detection of Carcinoembryonic Antigen. Nature 2019, 9, 7214. [Google Scholar] [CrossRef]
- Chowdhury, A.D.; Takemura, K.; Li, T.-C.; Suzuki, T.; Park, E.Y. Electrical pulse-induced electrochemical biosensor for hepatitis E virus detection. Nat. Commun. 2019, 10, 4–7. [Google Scholar] [CrossRef] [Green Version]
- Ye, W.; Guo, J.; Bao, X.; Chen, T.; Weng, W.; Chen, S.; Yang, M. Rapid and sensitive detection of bacteria response to antibiotics using nanoporous membrane and graphene quantum dot (GQDs)-based electrochemical biosensors. Materials 2017, 10, 603. [Google Scholar] [CrossRef] [Green Version]
- Mythili, S.; Malathi, N. Diagnostic markers of acute myocardial infarction. Biomed. Rep. 2015, 3, 743–748. [Google Scholar] [CrossRef] [Green Version]
- Nielsen, P.B.; Larsen, T.B.; Gorst-Rasmussen, A.; Skjøth, F.; Lip, G.Y.H. β-Blockers in atrial fibrillation patients with or without heart failure: Association with mortality in a nationwide cohort study. Circ. Hear. Fail. 2016, 9, 1–9. [Google Scholar] [CrossRef]
- Galea, R.; Cardillo, M.T.; Caroli, A.; Marini, M.G.; Sonnino, C.; Narducci, M.L.; Biasucci, L.M. Inflammation and C-reactive protein in atrial fibrillation: Cause or effect? Texas Hear. Inst. J. 2014, 41, 461–468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwon, C.H.; Kang, J.G.; Lee, H.J.; Kim, N.H.; Sung, J.W.; Cheong, E.; Sung, K.C. C-reactive protein and risk of atrial fibrillation in East Asians. Europace 2017, 19, 1643–1649. [Google Scholar] [CrossRef] [PubMed]
- Bing, X.; Wang, G. Label free C-reactive protein detection based on an electrochemical sensor for clinical application. Int. J. Electrochem. Sci. 2017, 12, 6304–6314. [Google Scholar] [CrossRef]
- Altintas, Z.; Uludag, Y.; Gurbuz, Y.; Tothill, I.E. Surface plasmon resonance based immunosensor for the detection of the cancer biomarker carcinoembryonic antigen. Talanta 2011, 86, 377–383. [Google Scholar] [CrossRef] [Green Version]
- Masdor, N.A.; Altintas, Z.; Tothill, I.E. Surface plasmon resonance immunosensor for the detection of Campylobacter jejuni. Chemosensors 2017, 5, 16. [Google Scholar] [CrossRef]
- Altintas, Z.; France, B.; Ortiz, J.O.; Tothill, I.E. Computationally modelled receptors for drug monitoring using an optical based biomimetic SPR sensor. Sens. Actuators B Chem. 2016, 224, 726–737. [Google Scholar] [CrossRef]
- Altintas, Z. Surface plasmon resonance based sensor for the detection of glycopeptide antibiotics in milk using rationally designed nanoMIPs. Sci. Rep. 2018, 8, 1–12. [Google Scholar] [CrossRef]
- Uludag, Y.; Esen, E.; Kokturk, G.; Ozer, H.; Muhammad, T.; Olcer, Z.; Basegmez, H.I.O.; Simsek, S.; Barut, S.; Gok, M.Y.; et al. Lab-on-a-chip based biosensor for the real-time detection of aflatoxin. Talanta 2016, 160, 381–388. [Google Scholar] [CrossRef]
- Weng, X.; Neethirajan, S. A microfluidic biosensor using graphene oxide and aptamer-functionalized quantum dots for peanut allergen detection. Biosens. Bioelectron. 2016, 85, 649–656. [Google Scholar] [CrossRef]
- Masdor, N.A.; Altintas, Z.; Tothill, I.E. Sensitive detection of Campylobacter jejuni using nanoparticles enhanced QCM sensor. Biosens. Bioelectron. 2016, 78, 328–336. [Google Scholar] [CrossRef]
- Devadoss, A.; Sudhagar, P.; Terashima, C.; Nakata, K.; Fujishima, A. Photoelectrochemical biosensors: New insights into promising photoelectrodes and signal amplification strategies. J. Photochem. Photobiol. C Photochem. Rev. 2015, 24, 43–63. [Google Scholar] [CrossRef]
- Ge, L.; Liu, Q.; Hao, N.; Wang, K. Recent developments of photoelectrochemical biosensors for food analysis. J. Mater. Chem. B Mater. Biol. Med. 2019, 7, 7283. [Google Scholar] [CrossRef]
- Chen, J.; Liu, Y.; Zhao, G.C. A novel photoelectrochemical biosensor for tyrosinase and thrombin detection. Sensors 2016, 16, 135. [Google Scholar] [CrossRef] [Green Version]
- Xiong, E.; Yan, X.; Zhang, X.; Li, Y.; Yang, R.; Meng, L.; Chen, J. A new photoelectrochemical biosensor for ultrasensitive determination of nucleic acids based on a three-stage cascade signal amplification strategy. Analyst 2018, 143, 2799–2806. [Google Scholar] [CrossRef]
- Zang, Y.; Lei, J.; Ju, H. Principles and applications of photoelectrochemical sensing strategies based on biofunctionalized nanostructures. Biosens. Bioelectron. 2017, 96, 8–16. [Google Scholar] [CrossRef]
- Tu, W.; Wang, W.; Lei, J.; Deng, S.; Ju, H. Chemiluminescence excited photoelectrochemistry using graphene-quantum dots nanocomposite for biosensing. Chem. Commun. 2012, 48, 6535–6537. [Google Scholar] [CrossRef]
- Fan, D.; Bao, C.; Khan, M.S.; Wang, C.; Zhang, Y.; Liu, Q.; Zhang, X.; Wei, Q. A novel label-free photoelectrochemical sensor based on N,S-GQDs and CdS co-sensitized hierarchical Zn2SnO4 cube for detection of cardiac troponin I. Biosens. Bioelectron. 2018, 106, 14–20. [Google Scholar] [CrossRef]
- Tian, J.; Zhao, H.; Quan, X.; Zhang, Y.; Yu, H.; Chen, S. Fabrication of graphene quantum dots/silicon nanowires nanohybrids for photoelectrochemical detection of microcystin-LR. Sens. Actuators B Chem. 2014, 196, 532–538. [Google Scholar] [CrossRef]
- Ahmed, S.R.; Mogus, J.; Chand, R.; Nagy, E.; Neethirajan, S. Optoelectronic fowl adenovirus detection based on local electric field enhancement on graphene quantum dots and gold nanobundle hybrid. Biosens. Bioelectron. 2018, 103, 45–53. [Google Scholar] [CrossRef]
- Ge, S.; Lan, F.; Liang, L.; Ren, N.; Li, L.; Liu, H.; Yan, M.; Yu, J. Ultrasensitive Photoelectrochemical Biosensing of Cell Surface N-Glycan Expression Based on the Enhancement of Nanogold-Assembled Mesoporous Silica Amplified by Graphene Quantum Dots and Hybridization Chain Reaction. ACS Appl. Mater. Interfaces 2017, 9, 6670–6678. [Google Scholar] [CrossRef]
- Prado, T.M.; Carrico, A.; Cincotto, F.H.; Fatibello-Filho, O.; Moraes, F.C. Bismuth vanadate/graphene quantum dot: A new nanocomposite for photoelectrochemical determination of dopamine. Sens. Actuators B Chem. 2019, 285, 248–253. [Google Scholar] [CrossRef]
- Yan, Y.; Liu, Q.; Du, X.; Qian, J.; Mao, H.; Wang, K. Visible light photoelectrochemical sensor for ultrasensitive determination of dopamine based on synergistic effect of graphene quantum dots and TiO2 nanoparticles. Anal. Chim. Acta 2015, 853, 258–264. [Google Scholar] [CrossRef]
- Wang, Y.; Zhou, Y.; Xu, L.; Han, Z.; Yin, H.; Ai, S. Photoelectrochemical apta-biosensor for zeatin detection based on graphene quantum dots improved photoactivity of graphite-like carbon nitride and streptavidin induced signal inhibition. Sens. Actuators B Chem. 2018, 257, 237–244. [Google Scholar] [CrossRef]
- Liu, Y.; Yan, K.; Okoth, O.K.; Zhang, J. A label-free photoelectrochemical aptasensor based on nitrogen-doped graphene quantum dots for chloramphenicol determination. Biosens. Bioelectron. 2015, 74, 1016–1021. [Google Scholar] [CrossRef]
- Ramdzan, N.S.M.; Fen, Y.W.; Omar, N.A.S.; Anas, N.A.A.; Daniyal, W.M.E.M.M.; Saleviter, S.; Zainudin, A.A. Optical and surface plasmon resonance sensing properties for chitosan/carboxyl-functionalized graphene quantum dots thin film. Optik (Stuttg.) 2019, 178, 802–812. [Google Scholar] [CrossRef]
- Sadrolhosseini, A.R.; Rashid, S.A.; Norhanisah, J.; Noor, A.S.; Isloor, A.M. Surface Plasmon Resonance Sensor Using Polypyrrole-Chitosan/Graphene Quantum Dots Layer for Detection of Sugar. Mater. Res. Express 2019, 6, 075028. [Google Scholar] [CrossRef]
- Wang, S.; Zhang, F.; Chen, C.; Cai, C. Ultrasensitive graphene quantum dots-based catalytic hairpin assembly amplification resonance light scattering assay for p53 mutant DNA detection. Sens. Actuators B Chem. 2019, 291, 42–47. [Google Scholar] [CrossRef]
- Wu, C.; Song, X.; Song, Z.; He, Z.; Dai, J. Ultrasensitive Detection of Protein Based on Graphene Quantum Dots with Resonance Light Scattering Technique. J. Nanosci. Nanotechnol. 2017, 18, 2680–2685. [Google Scholar] [CrossRef]
- Liu, F.; Ge, S.; Su, M.; Song, X.; Yan, M.; Yu, J. Electrochemiluminescence device for in-situ and accurate determination of CA153 at the MCF-7 cell surface based on graphene quantum dots loaded surface villous Au nanocage. Biosens. Bioelectron. 2015, 71, 286–293. [Google Scholar] [CrossRef]
- Li, L.; Zhang, Y.; Zhang, L.; Ge, S.; Liu, H.; Ren, N.; Yan, M.; Yu, J. Paper-Based Device for Colorimetric and Photoelectrochemical Quantification of the Flux of H2O2 Releasing from MCF-7 Cancer Cells. Anal. Chem. 2016, 88, 5369–5377. [Google Scholar] [CrossRef]
- Park, C.H.; Yang, H.; Lee, J.; Cho, H.H.; Kim, D.; Lee, D.C.; Kim, B.J. Multicolor Emitting Block Copolymer-Integrated Graphene Quantum Dots for Colorimetric, Simultaneous Sensing of Temperature, pH, and Metal Ions. Chem. Mater. 2015, 27, 5288–5294. [Google Scholar] [CrossRef]
- Lin, L.; Song, X.; Chen, Y.; Rong, M.; Zhao, T.; Wang, Y.; Jiang, Y.; Chen, X. Intrinsic peroxidase-like catalytic activity of nitrogen-doped graphene quantum dots and their application in the colorimetric detection of H2O2 and glucose. Anal. Chim. Acta 2015, 869, 89–95. [Google Scholar] [CrossRef]
- Chen, S.; Hai, X.; Chen, X.W.; Wang, J.H. In Situ growth of silver nanoparticles on graphene quantum dots for ultrasensitive colorimetric detection of H2O2 and glucose. Anal. Chem. 2014, 86, 6689–6694. [Google Scholar] [CrossRef] [PubMed]
- Nirala, N.R.; Abraham, S.; Kumar, V.; Bansal, A.; Srivastava, A.; Saxena, P.S. Colorimetric detection of cholesterol based on highly efficient peroxidase mimetic activity of graphene quantum dots. Sens. Actuators B Chem. 2015, 218, 42–50. [Google Scholar] [CrossRef]
































| Nanomaterials | Target Metal Ion | Linear Range | LOD | Reference |
|---|---|---|---|---|
| GQDs | Cu2+ | 0–15 µM | 0.226 µM | [116] |
| r–GQDs/GO | Pb2+ | 1–400 nM | 0.6 nM | [117] |
| GQDs@AuNPs | Pb2+ | 50 nM–4 µM | 16.7 nM | [118] |
| N,S–GQDs | Hg2+ | 0.1–15 µM | 0.14 nM | [119] |
| Val–GQDs | Hg2+ | 0.8 nM–1 µM | 0.4 nM | [120] |
| S–GQDs | Ag+ | 0.1–130 µM | 30 nM | [121] |
| GQDs@OPD | Ag+ | 0–115.2 µM | 250 nM | [122] |
| GQDs@AgNPs | Ag+ | 0–100 nM | 3.5 nM | [123] |
| Cit-UCNPs/GQDs | Ag+ | 2 × 10−4–1 µM | 60 pM | [124] |
| N–GQDs | Fe3+ | 1–1945 µM | 90 nM | [125] |
| RBD–GQDs | Fe3+ | 0–1 µM | 0.02 nM | [126] |
| N–GQDs | Fe3+ | 1–500 µM | 1 µM | [127] |
| B–GQDs | Fe3+ | 50 nM–420 µM | 31.2 nM | [128] |
| DA–GQDs | Fe3+ | 20 nM–2 µM | 7.6 nM | [129] |
| S–GQDs | Fe3+ | 0.01–0.70 µM | 4.2 nM | [130] |
| Nanomaterials | Receptor Type | Receptor | Target Analyte | Linear Range | Detection Limit | Sample(s) | Reference |
|---|---|---|---|---|---|---|---|
| GQDs | Nucleic acid | G-quadruplex/hemin DNAzyme | Uric acid | 2–300 µM | 500 nM | Human serum; Urine | [143] |
| N–GQDs | MIP | PDA-imprinted polymer | Thiacloprid | 0.1–10 mg L−1 | 0.3 mg L−1 | Vegetables | [144] |
| GQDs | Enzyme | HRP | o-DHB; m-DHB; p-DHB | 0.5–250 nM; 1–120 nM; 0.5–90 nM | 2 × 10−10 mol L−1; 8 × 10−10 mol L−1; 3 × 10−10 mol L−1 | Rain water; tap water | [145] |
| GQDs/MoS2 nanosheets | Nucleic acid | Aptamer | EpCAM | 3–54 nM | 450 pM | Human serum | [146] |
| GQDs | MIP | Atropine-imprinted polymer | Atropine | 0.5–300 × 10−9 g mL−1 | 0.22 × 10−9 g mL−1 | Urine; Human blood | [147] |
| GQDs/Fe3O4 | Antibody | Anti-IgG | IgG | 2.0–2.0 × 103 casts mL−1 | 2.0 casts mL−1 | Urine | [148] |
| N–GQDs | Nucleic acid | ss-DNA | Bleomycin | 0.34–1300 nmol L−1 | 0.34 nmol L−1 | Human serum | [149] |
| GQDs | Enzyme | Urease | Urea | 0.1–100 mM | 0.01 mM | Human blood | [150] |
| GQDs | MIP | tetracycline-imprinted polymer | tetracycline | 1–104 µg L−1 | 1 µg L−1 | Milk | [151] |
| NH2–GQDs | Nucleic acid | G-quadruplex/hemin DNAzyme | Human telomere DNA | 0.2–50 pM | 25 fM | Buffer; Human blood | [152] |
| Silica–GQDs | MIP | 4-NP imprinted polymer | 4-NP | 0.02–3 µg mL−1 | 9 ng mL−1 | water | [153] |
| GQDs/p-ABA | Enzyme | Tyrosinase | Artemisinin | 0.1–55 µM | 33 nM | Pharmaceuticals | [154] |
| GQDs | MIP | Silica-MIP | Ornidazole | 0.75–30 µM | 0.24 µM | Human plasma | [155] |
| Electrode | Nanomaterials | Receptor Type | Receptor | Target Analyte | Specimen | Linear Concentration Range | Detection Limit | Reference |
|---|---|---|---|---|---|---|---|---|
| GCE | HM–S–GQDs | Nucleic acid | Hemin/G-quadruplex DNAzyme | p53-gene | Buffer | 100 fM–100 nM | 66 nM | [214] |
| GCE | GQDs | Oligosaccharide | β-CD | L-Tyr; D-Tyr | Human serum | 6–100 µM 0.1–1 mM | 6.07 × 10−9 M 1.03 × 10−7 M | [215] |
| GCE | N–GQDs@SiO2 NPs | Nucleic acid | Aptamer | OTA | Peanut sample | 1 × 10−3–10 ng mL−1 | 0.5 × 10−3 ng mL−1 | [216] |
| GCE | MoS2‒GQDs | MIP | MCPA imprinted polymer | MCPA | Lake water; Tap water; Oat samples | 10 pM L−1–0.1 µM L−1 | 5.5 pM L−1 | [217] |
| GE | GQDs | Nucleic acid | DTC-DNA probe | HCV- 1b | Buffer | 5 fM–100 pM | 0.45 fM | [218] |
| GCE | NH2–PTCA/Au@Fe3O4/GQDs | Nucleic acid | DNA | miRNA | Buffer; Hela, HK-2, LO2, 22Rvl cell lines | 2.5 fM–50 pM | 0.83 fM | [219] |
| GCE | AuNPs/HM-S‒GQDs | Antibody | Anti-CEA | CEA | Buffer | 0.02–80 × 10−9 g mL−1 | 0.01 × 10−9 g mL−1 | [220] |
| Pt. | B–GQDs@AuNPs | Nucleic acid | DNA | miRNA-20 a | Human serum | 0.1–1 × 104 pM | 0.1 pM | [221] |
| GCE | Agcore@Aushell NPs/GQDs | Nucleic acid | DNA | lnc-RNA | Human serum | 1 fM–5 nM | 0.3 fM | [222] |
| Electrode | Nanomaterials | Receptor Type | Receptor | Target Analyte | Detection Technique(s) | Specimen | Linear Range | Detection Limit | Reference |
|---|---|---|---|---|---|---|---|---|---|
| GCE | GQDs | Enzyme | Laccase | EP | CV | Pharmaceuticals | 1–120 µM | 83 nM | [292] |
| GCE | N–GQDs | Oligosaccharide | β-CD | Cholesterol | DPV | Buffer, human serum | 0.5–100 nM | 80 nM | [293] |
| GCE | GQDs | Nucleic acid | DNA probe | HBV | CV; DPV | Human serum | 10 –500 nM | 1 nM | [294] |
| GCE | C3N4NTs@GQDs | MIP | CHL-imprinted polymer | CHL | SWV | Waste water | 0.01–1 nM | 2 pM | [295] |
| SPGE | PAMAM/GQDs/AuNPs@MWCNTs | Antibody | Anti-tTG | tTG | CV; DPV | Buffer; human serum | 20 fg mL−1–2 µg mL−1 | 20 fg mL−1 | [76] |
| GCE | Ru(core)@Au(shell) | MIP | PEA-imprinted polymer | PEA | CV; DPV | Urine | 1 pM–1 nM | 0.2 pM | [296] |
| GCE | AuPd NWs/D‒GQDs–GMA | Nucleic acid | DNA probe | Ara h1 | DPV | Peanut milk | 1 × 10−22–1 × 10−17 M | 4 × 10−23 M | [297] |
| GCE | GQDs/PdNPs | MIP | CIT-imprinted polymer | CIT | CV; DPV | Chicken egg | 1–5 nM | 0.2 nM | [298] |
| GCE | GQDs | Enzyme | Uric oxidase | UA | CV | Human serum | 1–800 µM | 0.3 µM | [299] |
| SPCE | GQDs implanted with N-ABA | MIP | Ifsosfamide-imprinted polymer | IFO | DPV | Aq. Solution; Blood plasma; Urine; Pharmaceuticals | 0.25–121 ng mL−1 0.31–116 ng mL−1 0.28–110 ng mL−1 0.32–109 ng mL−1 | 0.08 ng mL−1 0.11 ng mL−1 0.10 ng mL−1 0.10 ng mL−1 | [300] |
| GCE | NH2–GQDs/PBP(electroactive label) | Nucleic acid | ss-ds-DNA probes | micro RNA-25 | DPV | Spiked human serum | 0.3 nM–1 µM | 95 pM | [301] |
| GCE | GQDs | Enzyme | Glucose oxidase | Glucose | CV; DPV | Buffer | 10 µM–3 mM | 1.35 µM | [16] |
| GCE | GQDS/hNiNs | MIP | BPS-imprinted polymer | BPS | DPV | Mineral water; extracted plastic solution | 0.1–50 µM | 0.03 µM | [302] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mansuriya, B.D.; Altintas, Z. Applications of Graphene Quantum Dots in Biomedical Sensors. Sensors 2020, 20, 1072. https://0-doi-org.brum.beds.ac.uk/10.3390/s20041072
Mansuriya BD, Altintas Z. Applications of Graphene Quantum Dots in Biomedical Sensors. Sensors. 2020; 20(4):1072. https://0-doi-org.brum.beds.ac.uk/10.3390/s20041072
Chicago/Turabian StyleMansuriya, Bhargav D., and Zeynep Altintas. 2020. "Applications of Graphene Quantum Dots in Biomedical Sensors" Sensors 20, no. 4: 1072. https://0-doi-org.brum.beds.ac.uk/10.3390/s20041072