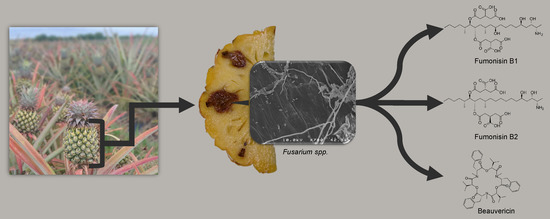
Diversity and Toxigenicity of Fungi that Cause Pineapple Fruitlet Core Rot
Abstract
:1. Introduction
2. Results
2.1. Morphological Identification
2.2. Distribution of Isolated Fungi
2.3. Phylogenetic Analysis
2.4. Mycotoxins Synthesized In Vitro
2.5. Mycotoxins in Pineapple Fruit
3. Discussion
4. Materials and Methods
4.1. Determination of Fungal Diversity
4.1.1. Fruit Sampling and Fungal Isolation
4.1.2. Morphological Comparisons
4.1.3. DNA Extraction, PCR, Sequencing, and Alignment
4.1.4. Phylogenetic Analysis
4.2. Toxigenic Potential
4.2.1. Pineapple Medium (PJA)
4.2.2. Optimal Medium for Mycotoxin Production
4.2.3. Culture Supplementation with Phenolic Acids and Enzymatic Complex
4.2.4. Inoculated and Naturally Infected Fruitlets
4.2.5. Mycotoxin Extraction and Quantification
4.3. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Tryon, H. Fruitlet core rot of pineapple. Qld. Agric. J. 1898, 3, 458–467. [Google Scholar]
- Simmonds, H.J. The work of the Pathological Branch. In Annual Report Queensland Department of Agriculture & Stock for the 1933-1934; CAB International: Wallingford, UK, 1934; pp. 67–70. [Google Scholar]
- Barker, H. Fruitlet black rot disease of pineapple. Phytopathology 1926, 16, 359. [Google Scholar]
- Serrano, G.B. Bacterial fruitlet rot of pineapple in the Phillipines. Phillipine J. Sci. 1928, 36, 271–305. [Google Scholar]
- Thompson, A. Pineapple fruit rots in Malaya. Malay Agric. J. 1937, 1937. [Google Scholar]
- Samson, R.A.; Yilmaz, N.; Houbraken, J.; Spierenburg, H.; Seifert, K.A.; Peterson, S.W.; Varga, J.; Frisvad, J.C. Phylogeny and nomenclature of the genus Talaromyces and taxa accommodated in Penicillium subgenus Biverticillium. Stud. Mycol. 2011, 70, 159–183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yilmaz, N.; Visagie, C.M.; Houbraken, J.; Frisvad, J.C.; Samson, R.A. Polyphasic taxonomy of the genus Talaromyces. Stud. Mycol. 2014, 78, 175–341. [Google Scholar] [CrossRef] [Green Version]
- Larsen, L.D. Disease in Pineapple; Hawaiian Sugar Planter’s Assoc.: Honolulu, HI, USA, 1910; p. 72. [Google Scholar]
- Aquije, G.M.D.V.; Zorzal, P.B.; Buss, D.S.; Ventura, J.A.; Fernandes, P.M.; Fernandes, A.A. Cell wall alterations in the leaves of fusariosis-resistant and susceptible pineapple cultivars. Plant Cell Rep. 2010, 29, 1109–1117. [Google Scholar] [CrossRef]
- Nirenberg, H.I.; O’Donnell, K. New Fusarium species and combinations within the Gibberella fujikuroi species complex. Mycologia 1998, 90, 434–458. [Google Scholar] [CrossRef]
- Aquije, G.M.D.V.; Korresa, A.M.N.; Buss, D.S.; Ventura, J.A.; Fernandes, P.M.B.; Fernandes, A.A.R. Effects of leaf scales of different pineapple cultivars on the epiphytic stage of Fusarium guttiforme. Crop Prot. 2011, 30, 375–378. [Google Scholar] [CrossRef] [Green Version]
- Kimati, H.; Tokeshi, H. Nota sobre a ocorrência de Fusarium causando resinose em abacaxi. Rev. Agric. 1964, 39, 131–133. [Google Scholar]
- Santos, C.; Ventura, J.A.; Lima, N. New Insights for Diagnosis of Pineapple Fusariosis by MALDI-TOF MS Technique. Curr. Microbiol. 2016, 73, 206–213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manicom, B.; Rabie, E.; Tustin, H. FURTHER INVESTIGATION OF THE EFFECTS OF THIOFLO ON BLACK SPOT OF PINEAPPLES. Acta Hortic. 2006, 157–162. [Google Scholar] [CrossRef]
- Petty, G.; Tustin, H.; Dicks, H. CONTROL OF BLACK SPOT DISEASE/FRUITLET CORE ROT IN QUEEN PINEAPPLE WITH INTEGRATED MEALYBUG, PINEAPPLE FRUIT MITE AND FUNGUS CONTROL PROGRAMMES. Acta Hortic. 2006, 143–149. [Google Scholar] [CrossRef]
- Jacobs, A.; Van Wyk, P.S.; Marasas, W.F.; Wingfield, B.D.; Wingfield, M.J.; Coutinho, T. Fusarium ananatum sp. nov. in the Gibberella fujikuroi species complex from pineapples with fruit rot in South Africa. Fungal Boil. 2010, 114, 515–527. [Google Scholar] [CrossRef] [Green Version]
- Stępień, Ł.; Koczyk, G.; Waśkiewicz, A. Diversity of Fusarium species and mycotoxins contaminating pineapple. J. Appl. Genet. 2013, 54, 367–380. [Google Scholar] [CrossRef] [Green Version]
- Gu, H.; Zhan, R.; Zhang, L.; Gong, D.; Jia, Z. First Report of Fusarium ananatum Causing Pineapple Fruitlet Core Rot in China. Plant Dis. 2015, 99, 1653–1653. [Google Scholar] [CrossRef]
- Jurado, M.; Marín, P.; Callejas, C.; Moretti, A.; Vázquez, C.; González-Jaén, M.T. Genetic variability and Fumonisin production by Fusarium proliferatum. Food Microbiol. 2010, 27, 50–57. [Google Scholar] [CrossRef]
- Kristensen, R.; Torp, M.; Kosiak, B.; Holst-Jensen, A. Phylogeny and toxigenic potential is correlated in Fusarium species as revealed by partial translation elongation factor 1 alpha gene sequences. Mycol. Res. 2005, 109, 173–186. [Google Scholar] [CrossRef]
- Stępień, Ł.; Koczyk, G.; Waśkiewicz, A. Genetic and phenotypic variation of Fusarium proliferatum isolates from different host species. J. Appl. Genet. 2011, 52, 487. [Google Scholar] [CrossRef] [Green Version]
- Johnson, M.O. The Pineapple; Paradise of the Pacific Press: Honolulu, HI, USA, 1935; p. 300. [Google Scholar]
- Pole-Evans, I.B. Report No. VI. Botany and Plant Pathology. Brown spot in pineapples. J. Dep. Agric. 1924, 9, 544–546. [Google Scholar]
- Edmonstone-Sammons, C. The Fungal Flora Associated with Black Spot of Pineapples. Some Aspects of the Microflora of Citrus Soils; Rhodes University: Grahamstown, South Africa, 1955. [Google Scholar]
- Oxenham, B. Etiology of fruitlet core rot of pineapple in Queensland. Qld. J. Agric. Sci. 1962, 19, 27–31. [Google Scholar]
- Rohrbach, K.G.; Namba, R.; Taniguchi, G. Endosulfan for Control of Pineapple Inter-fruitlet Corking, Leathery Pocket and Fruitlet Core Rot; The American Phytopathological Society: Saint Paul, MN, USA, 1981; pp. 1006–1007. [Google Scholar]
- Guerout, R. Les taches noires de l’ananas. Fruits 1974, 29, 489–499. [Google Scholar]
- Py, C.; Lacoeuilhe, J.-J.; Teisson, C. L’ananas: Sa Culture, Ses Produits; Maisonneuve et Larose: Paris, France, 1984; p. 562. [Google Scholar]
- Nguyen, P.-A.; Strub, C.; Fontana, A.; Schorr-Galindo, S. Crop molds and mycotoxins: Alternative management using biocontrol. Biol. Control 2017, 104, 10–27. [Google Scholar] [CrossRef]
- Mourichon, X. Contribution à l’etude des taches noires (Fruitlet core rot) et leathery pocket de l’ananas causés par Penicillium funiculosum Thom. en Côte d’Ivoire. Fruits 1983, 38, 209–224. [Google Scholar]
- Rohrbach, K.G.; Schmitt, D.; Ploetz, R.C. Diseases of pineapple. In Diseases of Tropical Fruit Crops; CAB International: Wallingford, UK, 2003; pp. 443–464. [Google Scholar]
- Vismer, H.F.; Sydenham, E.W.; Schlechter, M.; Brown, N.L.; Hocking, A.D.; Rheeder, J.P.; Marasas, W.F.O. Patulin-producing Penicillium species isolated from naturally infected apples in South Africa. S. Afr. J. Sci. 1996, 92, 530–534. [Google Scholar]
- Samson, R.A.; Houbraken, J.; Thrane, U.; Frisvad, J.C.; Andersen, B. Food and Indoor Fungi; Westerdijk Fungal Biodiversity Institute: Utrecht, The Netherlands, 2019; Volume 2, p. 481. [Google Scholar]
- Yassin, M.A.; El-Samawaty, A.-R.; Bahkali, A.; Moslem, M.; Abd-Elsalam, K.A.; Hyde, K.D. Mycotoxin-producing fungi occurring in sorghum grains from Saudi Arabia. Fungal Divers. 2010, 44, 45–52. [Google Scholar] [CrossRef]
- Avallone, S.; Guiraud, J.P.; Brillouet, J.M.; Teisson, C. Enzymatic browning and biochemical alterations in black spots of pineapple [Ananas comosus (L.) Merr.]. Curr. Microbiol. 2003, 47, 113–118. [Google Scholar] [CrossRef]
- Smith, B.G.; Harris, P.J. Ferulic acid is esterified to glucuronoarabinoxylans in pineapple cell walls. Phytochemistry 2001, 56, 513–519. [Google Scholar] [CrossRef]
- Steingass, C.B.; Glock, M.; Schweiggert, R.; Carle, R. Studies into the phenolic patterns of different tissues of pineapple (Ananas comosus [L.] Merr.) infructescence by HPLC-DAD-ESI-MSn and GC-MS analysis. Anal. Bioanal. Chem. 2015, 1–17. [Google Scholar] [CrossRef]
- Barral, B.; Chillet, M.; Léchaudel, M.; Lugan, R.; Schorr-Galindo, S. Coumaroyl-isocitric and caffeoyl-isocitric acids as markers of pineapple fruitlet core rot disease. Fruits 2019, 74, 11–17. [Google Scholar] [CrossRef]
- Barral, B.; Chillet, M.; Léchaudel, M.; Lartaud, M.; Verdeil, J.-L.; Conéjéro, G.; Schorr-Galindo, S. An Imaging Approach to Identify Mechanisms of Resistance to Pineapple Fruitlet Core Rot. Front. Plant Sci. 2019, 10, 1065. [Google Scholar] [CrossRef]
- Barral, B.; Chillet, M.; Minier, J.; Léchaudel, M.; Schorr-Galindo, S. Evaluating the response to Fusarium ananatum inoculation and antifungal activity of phenolic acids in pineapple. Fungal Biol. 2017, 121, 1045–1053. [Google Scholar] [CrossRef] [PubMed]
- Boutigny, A.L.; Atanasova-Penichon, V.; Benet, M.; Barreau, C.; Richard-Forget, F. Natural phenolic acids from wheat bran inhibit Fusarium culmorum trichothecene biosynthesis in vitro by repressing Tri gene expression. Eur. J. Plant Pathol. 2010, 127, 275–286. [Google Scholar] [CrossRef]
- Atanasova-Penichon, V.; Barreau, C.; Richard-Forget, F. Antioxidant secondary metabolites in cereals: Potential involvement in resistance to Fusarium and mycotoxin accumulation. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef] [Green Version]
- Gauthier, L.; Atanasova-Penichon, V.; Chéreau, S.; Richard-Forget, F. Metabolomics to decipher the chemical defense of cereals against Fusarium graminearum and deoxynivalenol accumulation. Int. J. Mol. Sci. 2015, 16, 24839–24872. [Google Scholar] [CrossRef]
- Forrer, H.; Musa, T.; Schwab, F.; Jenny, E.; Bucheli, T.; Wettstein, F.; Vogelgsang, S. Fusarium Head Blight Control and Prevention of Mycotoxin Contamination in Wheat with Botanicals and Tannic Acid. Toxins 2014, 6, 830–849. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ponts, N.; Pinson-Gadais, L.; Boutigny, A.-L.; Barreau, C.; Richard-Forget, F. Cinnamic-derived acids significantly affect Fusarium graminearum growth and in vitro synthesis of type B trichothecenes. Phytopathology 2011, 101, 929–934. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar]
- Zhang, N.; O’Donnell, K.; Sutton, D.A.; Nalim, F.A.; Summerbell, R.C.; Padhye, A.A.; Geiser, D.M. Members of the Fusarium solani species complex that cause infections in both humans and plants are common in the environment. J. Clin. Microbiol. 2006, 44, 2186–2190. [Google Scholar] [CrossRef] [Green Version]
- Sandoval-Denis, M.; Crous, P. Removing chaos from confusion: Assigning names to common human and animal pathogens in Neocosmospora. Persoonia 2018, 41, 109–129. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, K.; Humber, R.A.; Geiser, D.M.; Kang, S.; Park, B.; Robert, V.A.; Crous, P.W.; Johnston, P.R.; Aoki, T.; Rooney, A.P. Phylogenetic diversity of insecticolous fusaria inferred from multilocus DNA sequence data and their molecular identification via FUSARIUM-ID and Fusarium MLST. Mycologia 2012, 104, 427–445. [Google Scholar] [PubMed] [Green Version]
- Zhang, H.; Brankovics, B.; Luo, W.; Xu, J.; Xu, J.; Guo, C.; Guo, J.; Jin, S.; Chen, W.; Feng, J. Crops are a main driver for species diversity and the toxigenic potential of Fusarium isolates in maize ears in China. World Mycotoxin J. 2016, 9, 701–715. [Google Scholar]
- Kwon, S.-I.; Dohlen, C.D.v.; Anderson, A.J. Gene sequence analysis of an opportunistic wheat pathogen, an isolate of Fusarium proliferatum. Can. J. Bot. 2001, 79, 1115–1121. [Google Scholar]
- O’Donnell, K.; Nirenberg, H.I.; Aoki, T.; Cigelnik, E. A multigene phylogeny of the Gibberella fujikuroi species complex: Detection of additional phylogenetically distinct species. Mycoscience 2000, 41, 61–78. [Google Scholar]
- Yilmaz, N.; Houbraken, J.; Hoekstra, E.S.; Frisvad, J.C.; Visagie, C.M.; Samson, R.A. Delimitation and characterisation of Talaromyces purpurogenus and related species. Mol. Phylogeny Evol. Fungi 2012, 29, 39–54. [Google Scholar] [CrossRef] [Green Version]
- Jimenez, M.; Mateo, J.; Hinojo, M.; Mateo, R. Sugars and amino acids as factors affecting the synthesis of fumonisins in liquid cultures by isolates of the Gibberella fujikuroi complex. Int. J. Food Microbiol. 2003, 89, 185–193. [Google Scholar] [CrossRef]
- Rohrbach, K.G.; Pfeiffer, J.B. Susceptibility of pineapple cultivars to fruit diseases incited by Penicillium funiculosum and Fusarium moniliforme. Phytopathology 1976, 66, 1386–1390. [Google Scholar]
- O’Donnell, K.; Cigelnik, E.; Nirenberg, H.I. Molecular systematics and phylogeography of the Gibberella fujikuroi species complex. Mycologia 1998, 465–493. [Google Scholar]
- Raper, K.B.; Thom, C. Manual of the Penicillia; Williams & Wilkins: Baltimore, MD, USA, 1949. [Google Scholar]
- Fournier, P.; Benneveau, A.; Hardy, C.; Chillet, M.; Léchaudel, M. A predictive model based on a pluviothermic index for leathery pocket and fruitlet core rot of pineapple cv.‘Queen’. Eur. J. Plant Pathol. 2015, 142, 449–460. [Google Scholar]
- Ferruz, E.; Atanasova-Penichon, V.; Bonnin-Verdal, M.; Marchegay, G.; Pinson-Gadais, L.; Ducos, C.; Loran, S.; Arino, A.; Barreau, C.; Richard-Forget, F. Effects of Phenolic Acids on the Growth and Production of T-2 and HT-2 Toxins by Fusarium langsethiae and F. sporotrichioides. Molecules 2016, 21. [Google Scholar] [CrossRef] [Green Version]
- Wu, H.; Wang, Y.; Bao, W.; Liu, D.; Raza, W.; Huang, Q.; Mao, Z.; Shen, Q. Responses of Fusarium oxysporum f. sp niveum to exogenously added sinapic acid in vitro. Biol. Fertil. Soils 2009, 45, 443–447. [Google Scholar] [CrossRef]
- Kulik, T.; Stuper-Szablewska, K.; Bilska, K.; Busko, M.; Ostrowska-Kolodziejczak, A.; Zaluski, D.; Perkowski, J. trans-Cinnamic and Chlorogenic Acids Affect the Secondary Metabolic Profiles and Ergosterol Biosynthesis by Fusarium culmorum and F-graminearum Sensu Stricto. Toxins 2017, 9. [Google Scholar] [CrossRef]
- Pagnussatt, F.A.; Del Ponte, E.M.; Garda-Buffon, J.; Badiale-Furlong, E. Inhibition of Fusarium graminearum growth and mycotoxin production by phenolic extract from Spirulina sp. Pestic. Biochem. Physiol. 2014, 108, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Leslie, J.F.; Summerell, B.A.; Bullock, S. The Fusarium Laboratory Manual; Wiley Online Library: Hoboken, NJ, USA, 2006; Volume 2. [Google Scholar]
- Nelson, P.M.; WFO Toussoun, T. Fusarium Species: An Illustred Manual for Identification; The Pennsylvania State University: Pennsylvania, PA, USA, 1983. [Google Scholar]
- Rayner, R.W. A Mycological Colour Chart; CAB International: Wallingford, UK, 1970. [Google Scholar]
- Carbone, I.; Kohn, L.M. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 1999, 553–556. [Google Scholar]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Thierer, T.; Ashton, B.; Meintjes, P.; Drummond, D. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar]
- McWilliam, H.; Li, W.; Uludag, M.; Squizzato, S.; Park, Y.M.; Buso, N.; Cowley, A.P.; Lopez, R. Analysis tool web services from the EMBL-EBI. Nucleic Acids Res. 2013, 41, 597–600. [Google Scholar]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar]
- Guindon, S.; Dufayard, J.-F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar]
- R Core Team R. A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019. [Google Scholar]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: Berlin/Heidelberg, Germany, 2016. [Google Scholar]
- Kuznetsova, A.; Brockhoff, P.; Christensen, R. lmerTest Package: Tests in Linear Mixed Effects Models. J. Stat. Softw. 2017, 82, 1–26. [Google Scholar]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Stat. Methodol. 1995, 57, 289–300. [Google Scholar]
- Hothorn, T.; Bretz, F.; Westfall, P. Simultaneous Inference in General Parametric Models. Biom. J. 2008, 50, 346–353. [Google Scholar] [CrossRef] [PubMed] [Green Version]






| Species | Collection No. | FB1 (mg kg⁻1) | FB2 (mg kg⁻1) | BEA (mg kg⁻1) |
|---|---|---|---|---|
| Fusarium ananatum | clp019 | 0.02 | 0.01 | 0 |
| clp079 | 0.1 | 0.24 | 0 | |
| clp082 | 0.01 | 0.02 | 0 | |
| clp091 | 0.12 | 0.24 | 0 | |
| clp096 | 0.01 | 0.03 | 0 | |
| clp103 | 0.12 | 0.24 | 0 | |
| clp116 | 0.01 | 0.05 | 0 | |
| clp119 | 0.03 | 0.03 | 0 | |
| clp136 | 0.13 | 0.13 | 0 | |
| Fusarium oxysporum | clp076 | 0 | 0 | 23.68 |
| clp105 | 0 | 0 | 36.92 | |
| Fusarium proliferatum | clp081 | 96.7 | 11.2 | 103.2 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barral, B.; Chillet, M.; Doizy, A.; Grassi, M.; Ragot, L.; Léchaudel, M.; Durand, N.; Rose, L.J.; Viljoen, A.; Schorr-Galindo, S. Diversity and Toxigenicity of Fungi that Cause Pineapple Fruitlet Core Rot. Toxins 2020, 12, 339. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins12050339
Barral B, Chillet M, Doizy A, Grassi M, Ragot L, Léchaudel M, Durand N, Rose LJ, Viljoen A, Schorr-Galindo S. Diversity and Toxigenicity of Fungi that Cause Pineapple Fruitlet Core Rot. Toxins. 2020; 12(5):339. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins12050339
Chicago/Turabian StyleBarral, Bastien, Marc Chillet, Anna Doizy, Maeva Grassi, Laetitia Ragot, Mathieu Léchaudel, Noel Durand, Lindy Joy Rose, Altus Viljoen, and Sabine Schorr-Galindo. 2020. "Diversity and Toxigenicity of Fungi that Cause Pineapple Fruitlet Core Rot" Toxins 12, no. 5: 339. https://0-doi-org.brum.beds.ac.uk/10.3390/toxins12050339