Molecular Mechanisms Governing “Hair-Trigger” Induction of Shiga Toxin-Encoding Prophages
Abstract
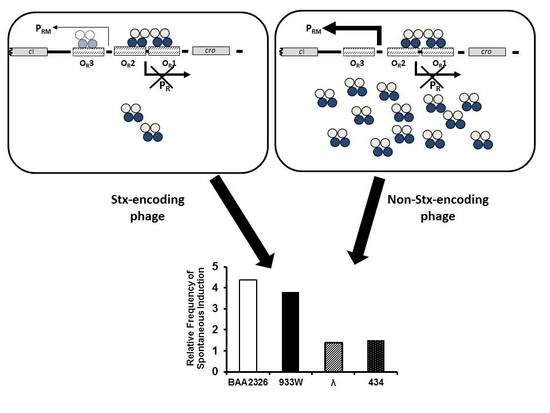
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Bacteriophages, and DNA
2.2. Quantitation of Intracellular Repressor Concentration
2.3. Quantitative Western Blots
2.4. Quantitative Dotblots
2.5. Purification of BAA2326 Repressor
2.6. Electrophoresis Mobility Shift Assays (EMSAs)
- OR15′GCGAATTCACTAAAGCACTTGCATTTAATAACTGTGCGTAATGATATGCCATAATAACTAAGAAGCTTCC3′
- OR25′GCGAATTCACTAAAGCACTTGCAATATGTAATGAAATATCTGCGGTGTTGACATTCCATAATAACTAAGAAGCTTCC3′
- OR35′GCGAATTCACTAAAGCACTTGCAATATCACCTTTGGTTATATGTAATGCCATAATAACTAAGAAGCTTCC3′
2.7. DNaseI Footprinting
2.8. Single Round In Vitro Transcription
2.9. Primer Extension Assay
3. Results
3.1. The BAA2326 Repressor Binds to Three Sites in the OR
- OR1- 5′ATAACTGTGCGTAATG3′
- OR2- 5′ATATCTGCGGTGTTGAC3′
- OR3- 5′CCTTTGGTTATATGTAAT3′
3.2. The BAA2326 Repressor Binds to OR1 and OR2 Cooperatively
3.3. Transcriptional Regulation at OR by BAA2326 Repressor
4. Discussion
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Ackermann, M.; Stecher, B.; Freed, N.E.; Songhet, P.; Hardt, W.-D.; Doebeli, M. Self-destructive cooperation mediated by phenotypic noise. Nature 2008, 454, 987–990. [Google Scholar] [CrossRef] [PubMed]
- Rangel, J.M.; Sparling, P.H.; Crowe, C.; Griffin, P.M.; Swerdlow, D.L. Epidemiology of Escherichia coli O157:H7 outbreaks, United States, 1982–2002. Emerg. Infect. Dis. 2005, 11, 603–609. [Google Scholar] [CrossRef] [PubMed]
- Luna-Gierke, R.; Griffin, P.; Gould, L.; Herman, K.; Bopp, C.; Strockbine, N.; Mody, R. Outbreaks of non-O157 Shiga toxin-producing Escherichia coli infection: USA. Epidemiol. Infect. 2014, 142, 2270–2280. [Google Scholar] [CrossRef] [PubMed]
- Trachtman, H.; Austin, C.; Lewinski, M.; Stahl, R.A.K. Renal and neurological involvement in typical Shiga toxin-associated HUS. Nat. Rev. Nephrol. 2012, 8, 658–669. [Google Scholar] [CrossRef] [PubMed]
- Mauro, S.A.; Koudelka, G.B. Shiga Toxin: Expression, Distribution, and Its Role in the Environment. Toxins 2011, 3, 608–625. [Google Scholar] [CrossRef] [PubMed]
- Waldor, M.K.; Friedman, D.I. Phage regulatory circuits and virulence gene expression. Curr. Opin. Microbiol. 2005, 8, 459–465. [Google Scholar] [CrossRef] [PubMed]
- Kimmitt, P.T.; Harwood, C.R.; Barer, M.R. Toxin gene expression by shiga toxin-producing Escherichia coli: The role of antibiotics and the bacterial SOS response. Emerg. Infect. Dis. 2000, 6, 458–465. [Google Scholar] [CrossRef] [PubMed]
- Newland, J.W.; Strockbine, N.A.; Miller, S.F.; O’Brien, A.D.; Holmes, R.K. Cloning of Shiga-like toxin structural genes from a toxin converting phage of Escherichia coli. Science 1985, 230, 179–181. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, A.D.; Newland, J.W.; Miller, S.F.; Holmes, R.K.; Smith, H.W.; Formal, S.B. Shiga-like toxin-converting phages from Escherichia coli strains that cause hemorrhagic colitis or infantile diarrhea. Science 1984, 226, 694–696. [Google Scholar] [CrossRef] [PubMed]
- Neely, M.N.; Friedman, D.I. Functional and genetic analysis of regulatory regions of coliphage H-19B: Location of shiga-like toxin and lysis genes suggest a role for phage functions in toxin release. Mol. Microbiol. 1998, 28, 1255–1267. [Google Scholar] [CrossRef] [PubMed]
- Neely, M.N.; Friedman, D.I. Arrangement and functional identification of genes in the regulatory region of lambdoid phage H-19B, a carrier of a Shiga-like toxin. Gene 1998, 223, 105–113. [Google Scholar] [CrossRef]
- Tiedtke, A.; Kiy, T.; Vosskuhler, C.; Rasmussen, L. Pathways of lysosomal enzyme secretion in Tetrahymena. Adv. Cell. Mol. Biol. Membr. 1993, 2A, 99–122. [Google Scholar]
- Shimizu, T.; Ohta, Y.; Noda, M. Shiga Toxin 2 Is Specifically Released from Bacterial Cells by Two Different Mechanisms. Infect. Immun. 2009, 77, 2813–2823. [Google Scholar] [CrossRef] [PubMed]
- Stolfa, G.; Koudelka, G.B. Entry and Killing of Tetrahymena thermophila by Bacterially Produced Shiga Toxin. mBio 2012, 4, e00416-12. [Google Scholar] [CrossRef] [PubMed]
- Arnold, J.W.; Koudelka, G.B. The Trojan Horse of the microbiological arms race: Phage-encoded toxins as a defence against eukaryotic predators. Environ. Microbiol. 2014, 16, 454–466. [Google Scholar] [CrossRef] [PubMed]
- Rozanov, D.V.; D’Ari, R.; Sineoky, S.P. RecA-independent pathways of lambdoid prophage induction in Escherichia coli. J. Bacteriol. 1998, 180, 6306–6315. [Google Scholar] [PubMed]
- Imamovic, L.; Muniesa, M. Characterizing RecA-Independent Induction of Shiga toxin2-Encoding Phages by EDTA Treatment. PLoS ONE 2012, 7, e32393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shkilnyj, P.; Koudelka, G.B. Effect of Salt Shock on the Stability of l imm434 Lysogens. J. Bacteriol. 2007, 189, 3115–3123. [Google Scholar] [CrossRef] [PubMed]
- Anderson, J.E. Cocrystals of the DNA-binding domain of phage 434 repressor and a synthetic phage 434 operator. Ph.D. Thesis, Harvard University, Cambridge, MA, USA, 1984. [Google Scholar]
- Daniels, D.L.; Schroeder, J.L.; Szybalski, W.; Sanger, F.; Coulson, A.R.; Hong, G.F.; Hill, D.F.; Petersen, G.F.; Blattner, F.R.; Hendrix, R.W.; et al. Lambda II; Cold Spring Harbor Laboratory: Cold Spring Harbor, NY, USA, 1983; pp. 519–676. [Google Scholar]
- Crane, J.K.; Byrd, I.W.; Boedeker, E.C. Virulence Inhibition by Zinc in Shiga-Toxigenic Escherichia coli. Infect. Immun. 2011, 79, 1696–1705. [Google Scholar] [CrossRef] [PubMed]
- Crane, J.K.; Broome, J.E.; Reddinger, R.M.; Werth, B.B. Zinc protects against shiga-toxigenic Escherichia coli by acting on host tissues as well as on bacteria. BMC Microbiol. 2014, 14, 145. [Google Scholar] [CrossRef] [PubMed]
- Livny, J.; Friedman, D.I. Characterizing spontaneous induction of Stx encoding phages using a selectable reporter system. Mol. Microbiol. 2004, 51, 1691–1704. [Google Scholar] [CrossRef] [PubMed]
- Wagner, P.L.; Livny, J.; Neely, M.N.; Acheson, D.W.; Friedman, D.I.; Waldor, M.K. Bacteriophage control of Shiga toxin 1 production and release by Escherichia coli. Mol. Microbiol. 2002, 44, 957–970. [Google Scholar] [CrossRef] [PubMed]
- Riley, L.W.; Remis, R.S.; Helgerson, S.D.; McGee, H.B.; Wells, J.G.; Davis, B.R.; Hebert, R.J.; Olcott, E.S.; Johnson, L.M.; Hargrett, N.T.; et al. Hemorrhagic Colitis Associated with A Rare Escherichia coli Serotype. N. Engl. J. Med. 1983, 308, 681–685. [Google Scholar] [CrossRef] [PubMed]
- Loos, S.; Ahlenstiel, T.; Kranz, B.; Staude, H.; Pape, L.; Härtel, C.; Vester, U.; Buchtala, L.; Benz, K.; Hoppe, B.; et al. An Outbreak of Shiga Toxin–Producing Escherichia coli O104:H4 Hemolytic Uremic Syndrome in Germany: Presentation and Short-term Outcome in Children. Clin. Infect. Dis. 2012, 55, 753–759. [Google Scholar] [CrossRef] [PubMed]
- Colon, M.P.; Chakraborty, D.; Pevzner, Y.; Koudelka, G.B. Mechanisms that Determine the Differential Stability of Stx(+) and Stx(−) Lysogens. Toxins (Basel) 2016, 8, 96. [Google Scholar] [CrossRef] [PubMed]
- Bullwinkle, T.J.; Koudelka, G.B. The lysis-lysogeny decision of bacteriophage 933W: A 933W repressor-mediated long-distance loop has no role in regulating 933W PRM activity. J. Bacteriol. 2011, 193, 3313–3323. [Google Scholar] [CrossRef] [PubMed]
- Arber, W.L.; Enquist, B.; Hohn, N.E.; Murray, K. Lambda II; Hendrix, R.W., Stahl, J.W.R.F.W., Weisberg, R.A., Eds.; Cold Spring Harbor Laboratory: Cold Spring Harbor, NY, USA, 1983; pp. 433–466. [Google Scholar]
- Donner, A.L.; Carlson, P.A.; Koudelka, G.B. Dimerization specificity of P22 and 434 repressors is determined by multiple polypeptide segments. J. Bacteriol. 1997, 179, 1253–1261. [Google Scholar] [CrossRef] [PubMed]
- Koudelka, A.P.; Hufnagel, L.A.; Koudelka, G.B. Purification and characterization of the repressor of the shiga toxin-encoding bacteriophage 933W: DNA binding, gene regulation, and autocleavage. J. Bacteriol. 2004, 186, 7659–7669. [Google Scholar] [CrossRef] [PubMed]
- Johnson, A.D.; Pabo, C.O.; Sauer, R.T. Bacteriophage lambda repressor and cro protein: Interactions with operator DNA. Methods Enzymol. 1982, 65, 839–856. [Google Scholar]
- Anderson, J.E.; Ptashne, M.; Harrison, S.C. Co-crystals of the DNA-binding domain of phage 434 repressor and a synthetic 434 operator. Proc. Natl. Acad. Sci. USA 1984, 81, 1307–1311. [Google Scholar] [CrossRef] [PubMed]
- Maxam, A.M.; Gilbert, W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980, 65, 499–560. [Google Scholar] [PubMed]
- Xu, J.; Koudelka, G.B. DNA-based positive control mutants in the binding site sequence of 434 repressor. J. Biol. Chem. 1998, 273, 24165–24172. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, M.E.; Brown, T.A.; Trumpower, B.L. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 1990, 18, 3091–3092. [Google Scholar] [CrossRef] [PubMed]
- Beckett, D.; Koblan, K.S.; Ackers, G.K. Quantitative study of protein association at picomolar concentrations: The lambda phage cl repressor. Anal. Biochem. 1991, 196, 69–75. [Google Scholar] [CrossRef]
- Bell, A.C.; Koudelka, G.B. Operator sequence context influences amino acid-base-pair interactions in 434 repressor-operator complexes. J. Mol. Biol. 1993, 234, 542–553. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, A.; Rodgers, D.W.; Drottar, M.; Ptashne, M.; Harrison, S.C. Recognition of a DNA operator by the repressor of phage 434: A view at high resolution. Science 1988, 242, 899–907. [Google Scholar] [CrossRef] [PubMed]
- Koblan, K.S.; Ackers, G.K. Site-specific enthalpic regulation of DNA transcription at bacteriophage lambda OR. Biochemistry 1992, 31, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Johnson, A.D.; Meyer, B.J.; Ptashne, M. Interaction between DNA-bound repressors govern regulation by the lambda repressor. Proc. Natl. Acad. Sci. USA 1979, 76, 5061–5065. [Google Scholar] [CrossRef] [PubMed]
- Wharton, R.P.; Ptashne, M. An à-helix determines the DNA specificity of a repressor. Trends Biochem. Sci. 1986, 11, 71–73. [Google Scholar] [CrossRef]
- Meyer, B.J.; Maurer, R.; Ptashne, M. Gene regulation at the right operator (OR) of bacteriophage lambda. II. OR1, OR2, and OR3: Their roles in mediating the effects of repressor and cro. J. Mol. Biol. 1980, 139, 163–194. [Google Scholar] [CrossRef]
- Tyler, J.S.; Mills, M.J.; Friedman, D.I. The operator and early promoter region of the Shiga toxin type 2-encoding bacteriophage 933W and control of toxin expression. J. Bacteriol. 2004, 186, 7670–7679. [Google Scholar] [CrossRef] [PubMed]
- Bushman, F.D. The bacteriophage 434 right operator. Roles of OR1, OR2 and OR3. J. Mol. Biol. 1993, 230, 28–40. [Google Scholar] [CrossRef] [PubMed]
- Levine, A.; Bailone, A.; Devoret, R. Cellular levels of the prophage λ and 434 repressors. J. Mol. Biol. 1979, 131, 655–661. [Google Scholar] [CrossRef]
- Morelli, M.J.; Ten Wolde, P.R.; Allen, R.J. DNA looping provides stability and robustness to the bacteriophage lambda switch. Proc. Natl. Acad. Sci. USA 2009, 106, 8101–8106. [Google Scholar] [CrossRef] [PubMed]
- Dodd, I.B.; Shearwin, K.E.; Perkins, A.J.; Burr, T.; Hochschild, A.; Egan, J.B. Cooperativity in long-range gene regulation by the lambda CI repressor. Genes Dev. 2004, 18, 344–354. [Google Scholar] [CrossRef] [PubMed]
- Wang, I.N. Lysis timing and bacteriophage fitness. Genetics 2006, 172, 17–26. [Google Scholar] [CrossRef] [PubMed]
- Shao, Y.P.; Wang, I.N. Bacteriophage adsorption rate and optimal lysis time. Genetics 2008, 180, 471–482. [Google Scholar] [CrossRef] [PubMed]





| Relative KDs | ||||||||
|---|---|---|---|---|---|---|---|---|
| Phage | λ | 434 | 933W | BAA2326 | ||||
| Site | Separate Sites | Intact OR | Separate Sites | Intact OR | Separate Sites | Intact OR | Separate Sites | Intact OR |
| OR 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| OR 2 | 75 | 2 | 14 | 1 | 10.3 | 13.6 | 83.3 | 1.5 |
| OR 3 | 30 | 30 | 5.5 | 12 | 15 | 26.4 | 2.5 | 2 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chakraborty, D.; Clark, E.; Mauro, S.A.; Koudelka, G.B. Molecular Mechanisms Governing “Hair-Trigger” Induction of Shiga Toxin-Encoding Prophages. Viruses 2018, 10, 228. https://0-doi-org.brum.beds.ac.uk/10.3390/v10050228
Chakraborty D, Clark E, Mauro SA, Koudelka GB. Molecular Mechanisms Governing “Hair-Trigger” Induction of Shiga Toxin-Encoding Prophages. Viruses. 2018; 10(5):228. https://0-doi-org.brum.beds.ac.uk/10.3390/v10050228
Chicago/Turabian StyleChakraborty, Dolonchapa, Eric Clark, Steven A. Mauro, and Gerald B. Koudelka. 2018. "Molecular Mechanisms Governing “Hair-Trigger” Induction of Shiga Toxin-Encoding Prophages" Viruses 10, no. 5: 228. https://0-doi-org.brum.beds.ac.uk/10.3390/v10050228