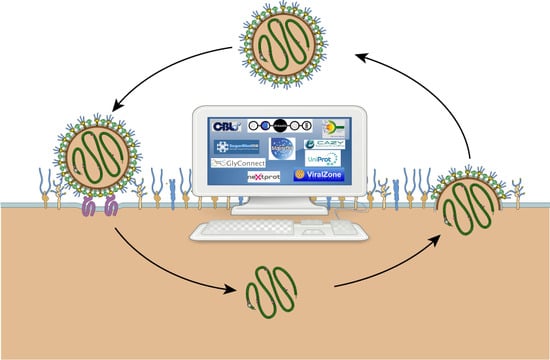
A Bioinformatics View of Glycan–Virus Interactions
Abstract
:1. Introduction
2. The Current Landscape of Bioinformatics Resources for Molecular Glycovirology
2.1. Overview
| Name | URL | Main Content | Relevance to Virology | Example of Use | Data Status | Created in | Reference |
|---|---|---|---|---|---|---|---|
| ViralZone | viralzone.expasy.org | Illustrated encyclopedia of viruses | Links textbook knowledge to sequence data | Explore the specific molecular biology of a virus | Curated | 2008 | [17] |
| UniProt | www.uniprot.org | Integrated knowledge of proteins | Provides protein sequence and functional information. | Find glycosylation sites in protein sequence | Curated (Swiss-Prot section) | 1986 | [18] |
| neXtprot | www.nextprot.org | Integrated knowledge of human proteins | Provides detailed functional information on human proteins | Characterize human host surface receptors | Partially curated | 2009 | [19] |
| PDBe | www.ebi.ac.uk/pdbe | 3D structures of proteins | Structural virology: capsid structures, host–virus interactions. | Examine the 3D structure of a capsid protein | Partially curated | 1980 | [29] |
| ChEBI | www.ebi.ac.uk/chebi | Information on chemical compounds | Describes chemical compounds interacting with viruses | Explore the molecule formula of a given compound | Curated | 2004 | [32] |
| PubChem | pubchem.ncbi.nlm.nih.gov | Information on chemical compounds and corresponding assays | Collects assays involving chemical compounds interacting with viruses | Find binding assays associated with a virus | Minimally curated | 2004 | [33] |
| CAZy | www.cazy.org | Carbohydrate Active Enzymes (CAZymes) | Families of viral CAZymes | Find viral CAZymes and associated Enzyme Nomenclature numbers | Curated | 1998 | [21] |
| GlyTouCan | www.glytoucan.org | Glycan 2D structures | Provides information on glycan structure. | Check existence of glycan bound by virus | Not curated | 2016 | [22] |
| Rhea | www.rhea.org | Enzymatic reactions | Describes virus specific enzyme reactions | Find substrate and product of CAZyme reaction | Curated | 2011 | [34] |
| MatrixDB | matrixdb.univ-lyon1.fr | Extracellular matrix components and interactions | Describes glycosaminoglycans and their interactions | Find interactions involving heparan sulfate | Curated | 2008 | [24] |
| SugarBind | sugarbind.expasy.org | Host glycans and pathogen lectins | Describes glycan–virus interactions | Explore pathogen lectins/adhesins binding a glycan motif | Curated | 2005 | [23] |
| UniLectin3D | www.unilectin.eu | Carbohydrate-binding proteins (not Ab) | Collects glycan–virus interactions | Inspect atomic details of glycan-haemagglutinin interactions | Curated | 2018 | [25] |
| GlyConnect | glyconnect.expasy.org | Integrated knowledge of glycoproteins | Describes glycoproteins | Find sialylated glycans on receptor proteins | Curated | 2017 | [20] |
| IMGT | www.imgt.org | Integrated knowledge of immunoglobulins | Collects antibody sequences and structures | Find antiviral antibody sequences | Curated | 1989 | [26] |
| IEDB | www.iedb.org | Collected knowledge of antigenic ligands (epitopes) | Collect experimental data characterizing the antigenicity of a virus. | Find viral antigenic peptides | Curated | 2003 | [27] |
| DAGR | ccr2.cancer.gov/resources/Cbl/Tools/Antibody | Carbohydrate-binding antibodies | Collects anti-glycan reagents | Find glycan binding motifs of antiviral antibodies | Curated | 2016 | [28] |
| Name | URL | Purpose | Applicable to Virology | Created in | Reference |
|---|---|---|---|---|---|
| LiteMol | www.litemol.org | Visualization of protein 3D structure | yes | 2015 | [35] |
| GlyS3 | glycoproteome.expasy.org/substructuresearch | Glycan substructure search | yes | 2015 | [36] |
| GlycoPattern | glycopattern.emory.edu | Analysis of glycan array data | yes | 2014 | [37] |
| GLAD | glycotoolkit.com/GLAD | Visualization and analysis of glycan array data | yes | 2019 | [38] |
| Glycome Atlas | rings.t.soka.ac.jp/GlycomeAtlasV5/index.html | Visualization of glycan array data | no | 2012 | [39] |

2.2. Illustration of Usage
3. Repertoire Mapping to Enrich Databases
3.1. Glycans as Virus Receptors in ViralZone

3.2. Emerging Knowledge in MatrixDB
3.3. SugarBindDB and UniLectin as Key Connectors

4. Discussion

5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Curry, F.E.; Adamson, R.H. Endothelial Glycocalyx: Permeability Barrier and Mechanosensor. Ann. Biomed. Eng. 2012, 40, 828–839. [Google Scholar] [CrossRef]
- Linden, S.K.; Sutton, P.; Karlsson, N.G.; Korolik, V.; McGuckin, M.A. Mucins in the mucosal barrier to infection. Mucosal Immunol. 2008, 1, 183–197. [Google Scholar] [CrossRef]
- Frey, A.; Giannasca, K.T.; Weltzin, R.; Giannasca, P.J.; Reggio, H.; Lencer, W.I.; Neutra, M.R. Role of the glycocalyx in regulating access of microparticles to apical plasma membranes of intestinal epithelial cells: Implications for microbial attachment and oral vaccine targeting. J. Exp. Med. 1996, 184, 1045–1059. [Google Scholar] [CrossRef]
- Stonebraker, J.R.; Wagner, D.; Lefensty, R.W.; Burns, K.; Gendler, S.J.; Bergelson, J.M.; Boucher, R.C.; O’Neal, W.K.; Pickles, R.J. Glycocalyx Restricts Adenoviral Vector Access to Apical Receptors Expressed on Respiratory Epithelium In Vitro and In Vivo: Role for Tethered Mucins as Barriers to Lumenal Infection. J. Virol. 2004, 78, 13755–13768. [Google Scholar] [CrossRef] [PubMed]
- Olofsson, S.; Bergström, T. Glycoconjugate glycans as viral receptors. Ann. Med. 2005, 37, 154–172. [Google Scholar] [CrossRef]
- Böhm, R.; Fleming, F.E.; Maggioni, A.; Dang, V.T.; Holloway, G.; Coulson, B.S.; von Itzstein, M.; Haselhorst, T. Revisiting the role of histo-blood group antigens in rotavirus host-cell invasion. Nat. Commun. 2015, 6, 5907. [Google Scholar] [CrossRef]
- Celerino da Silva, R.; Segat, L.; Crovella, S. Role of DC-SIGN and L-SIGN receptors in HIV-1 vertical transmission. Hum. Immunol. 2011, 72, 305–311. [Google Scholar] [CrossRef] [PubMed]
- Simmons, G.; Reeves, J.D.; Grogan, C.C.; Vandenberghe, L.H.; Baribaud, F.; Whitbeck, J.C.; Burke, E.; Buchmeier, M.J.; Soilleux, E.J.; Riley, J.L.; et al. DC-SIGN and DC-SIGNR Bind Ebola Glycoproteins and Enhance Infection of Macrophages and Endothelial Cells. Virology 2003, 305, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Lai, W.K.; Sun, P.J.; Zhang, J.; Jennings, A.; Lalor, P.F.; Hubscher, S.; McKeating, J.A.; Adams, D.H. Expression of DC-SIGN and DC-SIGNR on Human Sinusoidal Endothelium. Am. J. Pathol. 2006, 169, 200–208. [Google Scholar] [CrossRef] [PubMed]
- Londrigan, S.L.; Turville, S.G.; Tate, M.D.; Deng, Y.-M.; Brooks, A.G.; Reading, P.C. N-Linked Glycosylation Facilitates Sialic Acid-Independent Attachment and Entry of Influenza A Viruses into Cells Expressing DC-SIGN or L-SIGN. J. Virol. 2011, 85, 2990–3000. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.-Y.; Huang, Y.; Ganesh, L.; Leung, K.; Kong, W.-P.; Schwartz, O.; Subbarao, K.; Nabel, G.J. pH-Dependent Entry of Severe Acute Respiratory Syndrome Coronavirus Is Mediated by the Spike Glycoprotein and Enhanced by Dendritic Cell Transfer through DC-SIGN. J. Virol. 2004, 78, 5642–5650. [Google Scholar] [CrossRef]
- Bonomelli, C.; Doores, K.J.; Dunlop, D.C.; Thaney, V.; Dwek, R.A.; Burton, D.R.; Crispin, M.; Scanlan, C.N. The Glycan Shield of HIV Is Predominantly Oligomannose Independently of Production System or Viral Clade. PLoS ONE 2011, 6, e23521. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, Y.; Raghwani, J.; Allen, J.D.; Seabright, G.E.; Li, S.; Moser, F.; Huiskonen, J.T.; Strecker, T.; Bowden, T.A.; Crispin, M. Structure of the Lassa virus glycan shield provides a model for immunological resistance. Proc. Natl. Acad. Sci. USA 2018, 115, 7320–7325. [Google Scholar] [CrossRef]
- Shi, Q.; Wang, A.; Lu, Z.; Qin, C.; Hu, J.; Yin, J. Overview on the antiviral activities and mechanisms of marine polysaccharides from seaweeds. Carbohydr. Res. 2017, 453–454, 1–9. [Google Scholar] [CrossRef]
- Greene, A.C.; Giffin, K.A.; Greene, C.S.; Moore, J.H. Adapting bioinformatics curricula for big data. Brief. Bioinform. 2016, 17, 43–50. [Google Scholar] [CrossRef]
- Bagdonaite, I.; Wandall, H.H. Global aspects of viral glycosylation. Glycobiology 2018, 28, 443–467. [Google Scholar] [CrossRef] [PubMed]
- Masson, P.; Hulo, C.; De Castro, E.; Bitter, H.; Gruenbaum, L.; Essioux, L.; Bougueleret, L.; Xenarios, I.; Le Mercier, P. ViralZone: Recent updates to the virus knowledge resource. Nucleic Acids Res. 2013, 41, D579–D583. [Google Scholar] [CrossRef] [PubMed]
- UniProt Consortium. UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2018, 46, 2699. [Google Scholar] [CrossRef] [PubMed]
- Duek, P.; Gateau, A.; Bairoch, A.; Lane, L. Exploring the Uncharacterized Human Proteome Using neXtProt. J. Proteome Res. 2018, 17, 4211–4226. [Google Scholar] [CrossRef]
- Alocci, D.; Mariethoz, J.; Gastaldello, A.; Gasteiger, E.; Karlsson, N.G.; Kolarich, D.; Packer, N.H.; Lisacek, F. GlyConnect: Glycoproteomics Goes Visual, Interactive, and Analytical. J. Proteome Res. 2019, 18, 664–677. [Google Scholar] [CrossRef] [PubMed]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, D490–D495. [Google Scholar] [CrossRef] [PubMed]
- Tiemeyer, M.; Aoki, K.; Paulson, J.; Cummings, R.D.; York, W.S.; Karlsson, N.G.; Lisacek, F.; Packer, N.H.; Campbell, M.P.; Aoki, N.P.; et al. GlyTouCan: An accessible glycan structure repository. Glycobiology 2017, 27, 915–919. [Google Scholar] [CrossRef]
- Mariethoz, J.; Khatib, K.; Alocci, D.; Campbell, M.P.; Karlsson, N.G.; Packer, N.H.; Mullen, E.H.; Lisacek, F. SugarBindDB, a resource of glycan-mediated host–pathogen interactions. Nucleic Acids Res. 2016, 44, D1243–D1250. [Google Scholar] [CrossRef]
- Clerc, O.; Deniaud, M.; Vallet, S.D.; Naba, A.; Rivet, A.; Perez, S.; Thierry-Mieg, N.; Ricard-Blum, S. MatrixDB: Integration of new data with a focus on glycosaminoglycan interactions. Nucleic Acids Res. 2019, 47, D376–D381. [Google Scholar] [CrossRef]
- Bonnardel, F.; Mariethoz, J.; Salentin, S.; Robin, X.; Schroeder, M.; Perez, S.; Lisacek, F.; Imberty, A. UniLectin3D, a database of carbohydrate binding proteins with curated information on 3D structures and interacting ligands. Nucleic Acids Res. 2019, 47, D1236–D1244. [Google Scholar] [CrossRef]
- Lefranc, M.-P.; Giudicelli, V.; Duroux, P.; Jabado-Michaloud, J.; Folch, G.; Aouinti, S.; Carillon, E.; Duvergey, H.; Houles, A.; Paysan-Lafosse, T.; et al. IMGT®, the international ImMunoGeneTics information system® 25 years on. Nucleic Acids Res. 2015, 43, D413–D422. [Google Scholar] [CrossRef] [PubMed]
- Vita, R.; Mahajan, S.; Overton, J.A.; Dhanda, S.K.; Martini, S.; Cantrell, J.R.; Wheeler, D.K.; Sette, A.; Peters, B. The Immune Epitope Database (IEDB): 2018 update. Nucleic Acids Res. 2019, 47, D339–D343. [Google Scholar] [CrossRef]
- Sterner, E.; Flanagan, N.; Gildersleeve, J.C. Perspectives on Anti-Glycan Antibodies Gleaned from Development of a Community Resource Database. ACS Chem. Biol. 2016, 11, 1773–1783. [Google Scholar] [CrossRef] [PubMed]
- wwPDB Consortium; Burley, S.K.; Berman, H.M.; Bhikadiya, C.; Bi, C.; Chen, L.; Costanzo, L.D.; Christie, C.; Duarte, J.M.; Dutta, S.; et al. Protein Data Bank: The single global archive for 3D macromolecular structure data. Nucleic Acids Res. 2019, 47, D520–D528. [Google Scholar] [CrossRef]
- Sayers, E.W.; Barrett, T.; Benson, D.A.; Bolton, E.; Bryant, S.H.; Canese, K.; Chetvernin, V.; Church, D.M.; DiCuccio, M.; Federhen, S.; et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2011, 39, D38–D51. [Google Scholar] [CrossRef] [PubMed]
- Huang, P.; Farkas, T.; Marionneau, S.; Zhong, W.; Ruvoën-Clouet, N.; Morrow, A.L.; Altaye, M.; Pickering, L.K.; Newburg, D.S.; LePendu, J.; et al. Noroviruses Bind to Human ABO, Lewis, and Secretor Histo–Blood Group Antigens: Identification of 4 Distinct Strain-Specific Patterns. J. Infect. Dis. 2003, 188, 19–31. [Google Scholar] [CrossRef]
- Hastings, J.; Owen, G.; Dekker, A.; Ennis, M.; Kale, N.; Muthukrishnan, V.; Turner, S.; Swainston, N.; Mendes, P.; Steinbeck, C. ChEBI in 2016: Improved services and an expanding collection of metabolites. Nucleic Acids Res. 2016, 44, D1214–D1219. [Google Scholar] [CrossRef]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem 2019 update: Improved access to chemical data. Nucleic Acids Res. 2019, 47, D1102–D1109. [Google Scholar] [CrossRef]
- Morgat, A.; Lombardot, T.; Axelsen, K.B.; Aimo, L.; Niknejad, A.; Hyka-Nouspikel, N.; Coudert, E.; Pozzato, M.; Pagni, M.; Moretti, S.; et al. Updates in Rhea—An expert curated resource of biochemical reactions. Nucleic Acids Res. 2017, 45, D415–D418. [Google Scholar] [CrossRef]
- Sehnal, D.; Deshpande, M.; Vařeková, R.S.; Mir, S.; Berka, K.; Midlik, A.; Pravda, L.; Velankar, S.; Koča, J. LiteMol suite: Interactive web-based visualization of large-scale macromolecular structure data. Nat. Methods 2017, 14, 1121–1122. [Google Scholar] [CrossRef] [PubMed]
- Alocci, D.; Mariethoz, J.; Horlacher, O.; Bolleman, J.T.; Campbell, M.P.; Lisacek, F. Property Graph vs RDF Triple Store: A Comparison on Glycan Substructure Search. PLoS ONE 2015, 10, e0144578. [Google Scholar] [CrossRef]
- Agravat, S.B.; Saltz, J.H.; Cummings, R.D.; Smith, D.F. GlycoPattern: A web platform for glycan array mining. Bioinformatics 2014, 30, 3417–3418. [Google Scholar] [CrossRef] [PubMed]
- Mehta, A.Y.; Cummings, R.D. GLAD: GLycan Array Dashboard, a visual analytics tool for glycan microarrays. Bioinformatics 2019. [Google Scholar] [CrossRef]
- Konishi, Y.; Aoki-Kinoshita, K.F. The GlycomeAtlas tool for visualizing and querying glycome data. Bioinformatics 2012, 28, 2849–2850. [Google Scholar] [CrossRef] [PubMed]
- Sehnal, D.; Grant, O.C. Rapidly Display Glycan Symbols in 3D Structures: 3D-SNFG in LiteMol. J. Proteome Res. 2019, 18, 770–774. [Google Scholar] [CrossRef] [PubMed]
- Varki, A.; Cummings, R.D.; Aebi, M.; Packer, N.H.; Seeberger, P.H.; Esko, J.D.; Stanley, P.; Hart, G.; Darvill, A.; Kinoshita, T.; et al. Symbol Nomenclature for Graphical Representations of Glycans. Glycobiology 2015, 25, 1323–1324. [Google Scholar] [CrossRef] [PubMed]
- Mariethoz, J.; Alocci, D.; Gastaldello, A.; Horlacher, O.; Gasteiger, E.; Rojas-Macias, M.; Karlsson, N.G.; Packer, N.H.; Lisacek, F. Glycomics@ExPASy: Bridging the Gap. Mol. Cell. Proteom. 2018, 17, 2164–2176. [Google Scholar] [CrossRef] [PubMed]
- Stevens, J.; Blixt, O.; Tumpey, T.M.; Taubenberger, J.K.; Paulson, J.C.; Wilson, I.A. Structure and Receptor Specificity of the Hemagglutinin from an H5N1 Influenza Virus. Science 2006, 312, 404–410. [Google Scholar] [CrossRef]
- Raman, R.; Venkataraman, M.; Ramakrishnan, S.; Lang, W.; Raguram, S.; Sasisekharan, R. Advancing glycomics: Implementation strategies at the Consortium for Functional Glycomics. Glycobiology 2006, 16, 82R–90R. [Google Scholar] [CrossRef] [PubMed]
- Jinno, A.; Park, P.W. Role of glycosaminoglycans in infectious disease. Methods Mol. Biol. 2015, 1229, 567–585. [Google Scholar] [PubMed]
- Aquino, R.S.; Park, P.W. Glycosaminoglycans and infection. Front. Biosci. 2016, 21, 1260–1277. [Google Scholar]
- Kim, S.Y.; Zhao, J.; Liu, X.; Fraser, K.; Lin, L.; Zhang, X.; Zhang, F.; Dordick, J.S.; Linhardt, R.J. Interaction of Zika Virus Envelope Protein with Glycosaminoglycans. Biochemistry 2017, 56, 1151–1162. [Google Scholar] [CrossRef] [PubMed]
- Connell, B.J.; Lortat-Jacob, H. Human Immunodeficiency Virus and Heparan Sulfate: From Attachment to Entry Inhibition. Front. Immunol. 2013, 4, 385. [Google Scholar] [CrossRef]
- Ori, A.; Wilkinson, M.C.; Fernig, D.G. A Systems Biology Approach for the Investigation of the Heparin/Heparan Sulfate Interactome. J. Biol. Chem. 2011, 286, 19892–19904. [Google Scholar] [CrossRef]
- Ricard-Blum, S.; Lisacek, F. Glycosaminoglycanomics: Where we are. Glycoconj. J. 2017, 34, 339–349. [Google Scholar] [CrossRef] [PubMed]
- Clerc, O.; Mariethoz, J.; Rivet, A.; Lisacek, F.; Pérez, S.; Ricard-Blum, S. A pipeline to translate glycosaminoglycan sequences into 3D models. Application to the exploration of glycosaminoglycan conformational space. Glycobiology 2019, 29, 36–44. [Google Scholar] [CrossRef]
- Shanker, S.; Czako, R.; Sankaran, B.; Atmar, R.L.; Estes, M.K.; Prasad, B.V.V. Structural Analysis of Determinants of Histo-Blood Group Antigen Binding Specificity in Genogroup I Noroviruses. J. Virol. 2014, 88, 6168–6180. [Google Scholar] [CrossRef] [PubMed]
- Campbell, M.P.; Ranzinger, R.; Lütteke, T.; Mariethoz, J.; Hayes, C.A.; Zhang, J.; Akune, Y.; Aoki-Kinoshita, K.F.; Damerell, D.; Carta, G.; et al. Toolboxes for a standardised and systematic study of glycans. BMC Bioinform. 2014, 15, S9. [Google Scholar] [CrossRef] [PubMed]
- Tsuchiya, S.; Yamada, I.; Aoki-Kinoshita, K.F. GlycanFormatConverter: A conversion tool for translating the complexities of glycans. Bioinformatics 2018. [Google Scholar] [CrossRef] [PubMed]
- Herget, S.; Ranzinger, R.; Maass, K.; Lieth, C.-W.V.D. GlycoCT—A unifying sequence format for carbohydrates. Carbohydr. Res. 2008, 343, 2162–2171. [Google Scholar] [CrossRef]
- Uhlen, M.; Fagerberg, L.; Hallstrom, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, A.; Kampf, C.; Sjostedt, E.; Asplund, A.; et al. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Ortega, V.; Stone, J.A.; Contreras, E.M.; Iorio, R.M.; Aguilar, H.C. Addicted to sugar: Roles of glycans in the order Mononegavirales. Glycobiology 2019, 29, 2–21. [Google Scholar] [CrossRef] [PubMed]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Le Mercier, P.; Mariethoz, J.; Lascano-Maillard, J.; Bonnardel, F.; Imberty, A.; Ricard-Blum, S.; Lisacek, F. A Bioinformatics View of Glycan–Virus Interactions. Viruses 2019, 11, 374. https://0-doi-org.brum.beds.ac.uk/10.3390/v11040374
Le Mercier P, Mariethoz J, Lascano-Maillard J, Bonnardel F, Imberty A, Ricard-Blum S, Lisacek F. A Bioinformatics View of Glycan–Virus Interactions. Viruses. 2019; 11(4):374. https://0-doi-org.brum.beds.ac.uk/10.3390/v11040374
Chicago/Turabian StyleLe Mercier, Philippe, Julien Mariethoz, Josefina Lascano-Maillard, François Bonnardel, Anne Imberty, Sylvie Ricard-Blum, and Frédérique Lisacek. 2019. "A Bioinformatics View of Glycan–Virus Interactions" Viruses 11, no. 4: 374. https://0-doi-org.brum.beds.ac.uk/10.3390/v11040374