Bacteriophage ZCSE2 is a Potent Antimicrobial against Salmonella enterica Serovars: Ultrastructure, Genomics and Efficacy
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Growth Media
2.2. Antibiotic Sensitivity Test
2.3. Bacteriophage Isolation, Amplification, and Purification
2.4. Lytic Profile of Isolated Phages
2.5. Efficiency of Plating
2.6. Phage Genome Size Determination Using (PFGE)
2.7. In Vitro ZCSE2 Lytic Activity
2.8. Bacteriophage Insensitive Mutant Frequency
2.9. ZCSE2 pH Stability
2.10. ZCSE2 Activity at 4 °C
2.11. ZCSE2 Genome Sequencing
2.12. Morphology Investigation by Transmission Electron Microscopy (TEM)
2.12.1. TEM Device Specification and Examination Conditions
2.12.2. TEM Sample Preparation
2.12.3. TEM Data Analysis
2.13. AFM Sample Preparation
2.13.1. AFM Device Specification
2.13.2. Post-Processing of AFM Images
2.14. Statistical Analysis
3. Results
3.1. Bacterial Sensitivity to Antibiotics
3.2. Lytic Profile and EOP of Isolated Bacteriophages
3.3. Antibacterial Efficacy of ZCSE2 In Vitro
3.4. Phage Genome Size Determination Using PFGE and Sequencing
3.5. ZCSE2 Complete Genome Sequence
3.6. ZCSE2 pH Stability
3.7. ZCSE2 Stability at Low Temperature
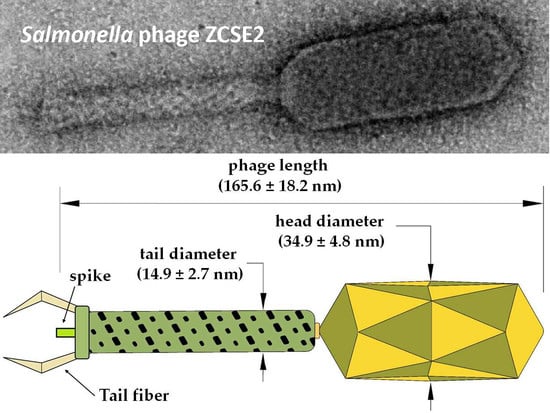
3.8. ZCSE2 Characterization and Imaging Bacterial Lysis with TEM
3.9. ZCSE2 Characterization and Imaging Bacterial Lysis with AFM
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Balasubramanian, R.; Im, J.; Lee, J.S.; Jeon, H.J.; Mogeni, O.D.; Kim, J.H.; Rakotozandrindrainy, R.; Baker, S.; Marks, F. The global burden and epidemiology of invasive non-typhoidal Salmonella infections. Hum. Vaccin. Immunother. 2019, 15, 1421–1426. [Google Scholar] [CrossRef] [PubMed]
- Kariuki, S.; Gordon, M.A.; Feasey, N.; Parry, C.M. Antimicrobial resistance and management of invasive Salmonella disease. Vaccine 2015, 33, C21–C29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hammarlöf, D.L.; Kröger, C.; Owen, S.V.; Canals, R.; Lacharme-Lora, L.; Wenner, N.; Schager, A.E.; Wells, T.J.; Henderson, I.R.; Wigley, P.; et al. Role of a single noncoding nucleotide in the evolution of an epidemic African clade of Salmonella. Proc. Natl. Acad. Sci. USA 2018, 115, E2614–E2623. [Google Scholar] [CrossRef] [Green Version]
- Canals, R.; Hammarlöf, D.L.; Kröger, C.; Owen, S.V.; Fong, W.Y.; Lacharme-Lora, L.; Zhu, X.; Wenner, N.; Carden, S.E.; Honeycutt, J.; et al. Adding function to the genome of African Salmonella Typhimurium ST313 strain D23580. PLoS Biol. 2019, 17, e3000059. [Google Scholar] [CrossRef] [Green Version]
- Ao, T.T.; Feasey, N.A.; Gordon, M.A.; Keddy, K.H.; Angulo, F.J.; Crump, J.A. Global burden of invasive nontyphoidal Salmonella disease, 2010. Emerg. Infect. Dis. 2015, 21. [Google Scholar] [CrossRef] [Green Version]
- Feasey, N.A.; Dougan, G.; Kingsley, R.A.; Heyderman, R.S.; Gordon, M.A. Invasive non-typhoidal Salmonella disease: An emerging and neglected tropical disease in Africa. Lancet 2012, 379, 2489–2499. [Google Scholar] [CrossRef]
- Crump, J.A.; Sjölund-Karlsson, M.; Gordon, M.A.; Parry, C.M. Epidemiology, Clinical presentation, laboratory diagnosis, antimicrobial resistance, and antimicrobial management of invasive Salmonella infections. Clin. Microbiol. Rev. 2015, 28, 901–937. [Google Scholar] [CrossRef] [Green Version]
- Hooton, S.P.; Atterbury, R.J.; Connerton, I.F. Application of a bacteriophage cocktail to reduce Salmonella Typhimurium U288 contamination on pig skin. Int. J. Food Microbiol. 2011, 151, 157–163. [Google Scholar] [CrossRef]
- Xu, Y.; Liu, Y.; Liu, Y.; Pei, J.; Yao, S.; Cheng, C. Bacteriophage therapy against Enterobacteriaceae. Virol. Sin. 2015, 30, 11–18. [Google Scholar] [CrossRef]
- Wernicki, A.; Nowaczek., A.; Urban-Chmiel, R. Bacteriophage therapy to combat bacterial infections in poultry. Virol. J. 2017, 14, 179. [Google Scholar] [CrossRef]
- Nagel, T.E.; Chan, B.K.; De Vos, D.; El-Shibiny, A.; Kang’ethe, E.K.; Makumi, A.; Pirnay, J.P. The developing world urgently needs phages to combat pathogenic bacteria. Front. Microbiol. 2016, 7, 882. [Google Scholar] [CrossRef] [PubMed]
- Grant, A.Q.; Parveen, S.; Schwarz, J.; Hashem, F.; Vimini, B. Reduction of Salmonella in ground chicken using a bacteriophage. Poult. Sci. 2017, 96, 2845–2852. [Google Scholar] [CrossRef] [PubMed]
- Weber-Dąbrowska, B.; Jończyk-Matysiak, E.; Żaczek, M.; Łobocka, M.; Łusiak-Szelachowska, M.; Górski, A. Bacteriophage Procurement for Therapeutic Purposes. Front. Microbiol. 2016, 7, 1177. [Google Scholar] [CrossRef]
- Dubrovin, E.V.; Popova, A.V.; Kraevskiy, S.V.; Ignatov, S.G.; Ignatyuk, T.E.; Yaminsky, I.V.; Volozhantsev, N.V. Atomic force microscopy analysis of the Acinetobacter baumannii bacteriophage AP22 lytic cycle. PLoS ONE 2012, 7, e47348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuznetsov, Y.G.; Malkin, A.; Lucas, R.; Plomp, M.; McPherson, A. Imaging of viruses by atomic force microscopy. J. Gen. Virol. 2001, 82, 2025–2034. [Google Scholar] [CrossRef] [PubMed]
- Marchetti, M.; Wuite, G.; Roos, W. Atomic force microscopy observation and characterization of single virions and virus-like particles by nano-indentation. Curr. Opin. Virol. 2016, 18, 82–88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alsteens, D.; Pesavento, E.; Cheuvart, G.; Dupres, V.; Trabelsi, H.; Soumillion, P.; Dufrêne, Y.F. Controlled manipulation of bacteriophages using single-virus force spectroscopy. ACS Nano 2009, 3, 3063–3068. [Google Scholar] [CrossRef]
- CSLI. Methods for Determining Bactericidal Activity of Antimicrobial Agents: Approved Guideline; CSLI Document M26-A; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 1999. [Google Scholar]
- Adams, M.H. Bacteriophages; Interscience Publishers Inc.: New York, NY, USA, 1959. [Google Scholar]
- Marcó, M.B.; Moineau, S.; Quiberoni, A. Bacteriophages and dairy fermentations. Bacteriophage 2012, 2, 149–158. [Google Scholar] [CrossRef] [Green Version]
- Mazzocco, A.; Waddell, T.E.; Lingohr, E.; Johnson, R.P. Enumeration of bacteriophages using the small drop plaque assay system. Methods Mol. Biol. 2009, 501, 81–85. [Google Scholar]
- Kutter, E. Phage host range and efficiency of plating. Methods Mol. Biol. 2009, 501, 141–149. [Google Scholar]
- Senczek, D.; Stephan, R.; Untermann, F. Pulsed-field gel electrophoresis (PFGE) typing of Listeria strains isolated from a meat processing plant over a 2-year period. Int. J. Food Microbiol. 2000, 62, 155–159. [Google Scholar] [CrossRef]
- Armon, R.; Kott, Y. A simple, rapid and sensitive presence/absence detection test for bacteriophage in drinking water. J. Appl. Bacteriol. 1993, 74, 490–496. [Google Scholar] [CrossRef] [PubMed]
- Miles, A.A.; Misra, S.S.; Irwin, J.O. The estimation of the bactericidal power of the blood. J. Hyg (Lond.) 1938, 38, 732–749. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Flynn, G.; Ross, R.P.; Fitzgerald, G.F.; Coffey, A. Evaluation of a cocktail of three bacteriophages for bio control of Escherichia coli O157:H7. Appl. Environ. Microbiol. 2004, 70, 3417–3424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arndt, D.; Grant, J.R.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. PHASTER: A better, faster version of the PHAST phage search tool. Nucl. Acids Res. 2016, 44, W16–W21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zimmermann, L.; Stephens, A.; Nam, S.Z.; Rau, D.; Kübler, J.; Lozajic, M.; Gabler, F.; Söding, J.; Lupas, A.N.; Alva, V. A completely reimplemented MPI bioinformatics toolkit with a new HHpred server at its core. J. Mol. Biol. 2018, 430, 2237–2239. [Google Scholar] [CrossRef] [PubMed]
- Nečas, D.; Klapetek, P. Gwyddion: An open-source software for SPM data analysis. Cent. Eur. J. Phys. 2012, 10, 181–188. [Google Scholar] [CrossRef]
- Besser, T.; Goldoft, M.; Pritchett, L.; Khakhria, R.; Hancock, D.; Rice, D.; Gay, J.; Johnson, W.; Gay, C. Multiresistant Salmonella Typhimurium DT104 infections of humans and domestic animals in the Pacific Northwest of the United States. Epidemiol. Infect. 2000, 124, 193–200. [Google Scholar] [CrossRef]
- Moye, Z.D.; Woolston, J.; Sulakvelidze, A. Bacteriophage applications for food production and processing. Viruses 2018, 10, 205. [Google Scholar] [CrossRef] [Green Version]
- Abedon, S.T. Lysis from without. Bacteriophage 2011, 1, 46–49. [Google Scholar] [CrossRef]
- Capparelli, R.; Nocerino, N.; Iannaccone, M.; Ercolini, D.; Parlato, M.; Chiara, M.; Iannelli, D. Bacteriophage therapy of Salmonella enterica: A fresh appraisal of bacteriophage therapy. Int J. Infect. Dis. 2010, 201, 52–61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oechslin, F. Resistance development to bacteriophages occurring during bacteriophage therapy. Viruses 2018, 10, 351. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.; Virk, S.M.; Shi, J.; Zhou, Y.; Willias, S.P.; Morsy, M.K.; Abdelnabby, H.E.; Liu, J.; Wang, X.; Li, J. Isolation, characterization, and application of bacteriophage LPSE1 against Salmonella enterica in ready to eat (RTE) foods. Front. Microbiol. 2018, 9, 1046. [Google Scholar] [CrossRef] [PubMed]
- Phongtang, W.; Choi, G.P.; Chukeatirote, E.; Ahn, J. Bacteriophage control of Salmonella Typhimurium in milk. Food Sci. Biotechnol. 2018, 28, 297–301. [Google Scholar] [CrossRef]






| Class | Antibiotics | Diameter of Inhibition Zone (cm) | Results |
|---|---|---|---|
| Cephalosporins Second generation | Cefoxitin | 2.2 | Intermediate |
| Cefaclor | 0.8 | Resistant | |
| Cephalosporins Third generation | Cefotaxime | 2.2 | Resistant |
| Chloramphenicol | 2.9 | Susceptible | |
| Ceftazidime | 2.3 | Susceptible | |
| Ceftriaxone | 3.1 | Susceptible | |
| Macrolides | Clarithromycin | 0.9 | Resistant |
| Erythromycin | 0.8 | Resistant | |
| Vancomycin | 0 | Resistant | |
| Azithromycin | 1.4 | Intermediate | |
| Quinolones | Levofloxacin | 3.4 | Susceptible |
| Ciprofloxacin | 3.0 | Intermediate | |
| Penicillins | Piperacillin | 2.5 | Susceptible |
| Oxacillin | 0 | Resistant | |
| Oxazolidinones | Linezolid | 0.8 | Resistant |
| Lincosamides | Clindamycin | 0 | Resistant |
| Nitrofurans | Nitrofurantoin | 1.3 | Resistant |
| Carbapenems | Ertapenem | 3.0 | Susceptible |
| Aminoglycosides | Amikacin | 1.9 | Susceptible |
| Aminocoumarin | Novobiocin | 1.2 | Resistant |
| Tetracyclines | Tetracycline | 2.8 | Susceptible |
| Salmonella Strains | ZCSE2 | ZCSE3 | ZCSE4 | ZCSE5 |
|---|---|---|---|---|
| S. Agama WT | + | + | + | − |
| S. Togla Amersham 5.8.95 | + | − | − | + |
| S. Amsterdam WT | + | − | − | − |
| S. Typhimurium DT104 NCTC 13348 | + | + | − | − |
| S. Atlanta NCTC 9986 | + | + | + | − |
| S. Typhimurium LT2 | + | + | − | − |
| S. Bareilly NCTC 5745 | + | + | − | − |
| S. Typhimurium WT Rawlings | + | + | − | − |
| S. Derby WT | + | + | − | − |
| S. Typhimurium S21344 | + | + | − | − |
| S. Enteritidis HOOO WT | + | + | − | − |
| S. Typhimurium WT Turner | + | + | − | − |
| S. Enteritidis WT (Platten) | + | + | − | − |
| S. Typhimurium U288 | + | + | − | − |
| S. Enteritidis SA029 | + | + | − | − |
| S. Virchow WT | + | + | + | − |
| S. Enteritidis WT Harrison | + | + | − | − |
| S. Hadar WT | + | + | + | − |
| S. Infantis NCTC 6903 | + | − | − | + |
| S. Kedougou | + | − | − | + |
| S. Kubacha WT | + | + | − | − |
| S. Montevideo NCTC 5797 | + | − | − | − |
| S. Montevideo WT | + | − | − | − |
| S. Senftenburg WT | − | − | − | − |
| S. Thompson NCTC 2252 | + | + | − | − |
| Protein ID | Chromosomal Loci (nt) | Putative Function |
|---|---|---|
| QBZ70504 | 1–978 | Major capsid protein |
| QBZ70505 | 1051–1425 | HNH endonuclease |
| QBZ70506 | 1455–1955 | DCTP pyrophosphatase |
| QBZ70507 | 1955–2455 | DNA primase |
| QBZ70508 | 2448–2825 | Head-to-tail interface protein |
| QBZ70510 | 3276–3896 | Minor tail fiber |
| QBZ70511 | 3911–6853 | Tail protein |
| QBZ70512 | 6923–7564 | Head-to-tail interface protein |
| QBZ70513 | 7577–9451 | Sheath structural protein |
| QBZ70514 | 9539–10,678 | Cell adhesion protein |
| QBZ70515 | 10,689–11,120 | DNA helicase |
| QBZ70516 | 11,137–11,565 | Membrane-associated initiation of head vertex |
| QBZ70517 | 11,616–11,750 | RecA-like recombination protein |
| QBZ70518 | 11,731–13,392 | DNA polymerase |
| QBZ70520 | 14,619–15,569 | RegA-like translational repressor |
| QBZ70521 | 15,559–16,203 | Baseplate assembly V |
| QBZ70522 | 16,212–16,583 | Clamp-loader subunit |
| QBZ70523 | 16,587–17,750 | Sliding clamp DNA polymerase |
| QBZ70524 | 17,743–18,396 | RNA polymerase binding protein |
| QBZ70525 | 18,389–19,738 | Tail fiber |
| QBZ70526 | 19,738–20,280 | Tail fiber assembly protein |
| QBZ70527 | 20,283–20,792 | Tail fiber chaperone |
| QBZ70528 | 20,894–21,157 | dsDNA binding protein |
| QBZ70529 | 21,201–21,677 | Lysozyme |
| QBZ70530 | 21,656–21,985 | DUF2570 domain-containing protein |
| QBZ70531 | 22,203–22,427c | TraR/DksA family transcriptional regulator |
| QBZ70533 | 22,711–24,681c | DNA polymerase 1 |
| QBZ70534 | 24,678–25,676c | DNA polymerase beta subunit |
| QBZ70535 | 25,709–26,593c | Thymidylate synthase |
| QBZ70536 | 26,593–27,186 | NTP pyrophosphohydrolase |
| QBZ70541 | 28,955–30,628c | ATP-RNA helicase |
| QBZ70543 | 30,835–31,710c | Cas4 family exonuclease |
| QBZ70544 | 31,712–32,176c | Deoxycytidylate deaminase |
| QBZ70545 | 32,237–32,776c | DUF669 domain-containing protein |
| QBZ70546 | 32,881–33,771c | σ factor for late transcription |
| QBZ70550 | 35,919–36,215c | Glyoxalase |
| QBZ70552 | 36,284–36,616c | Host-nuclease inhibitor protein |
| QBZ70558 | 38,983–39,315 | Glutaredoxin |
| QBZ70559 | 39,290–41,824 | Anaerobic NTP reductase small subunit |
| QBZ70560 | 42,319–42,747 | Protector of prophage-induced early lysis |
| QBZ70574 | 47,327–47,998 | Dihydrofolate reductase |
| QBZ70575 | 47,995–48,495 | Thymidylate kinase |
| QBZ70579 | 50,055–51,503 | Terminase large subunit |
| QBZ70580 | 51,505–53,067 | Portal protein |
| QBZ70581 | 53,246–53962 | Scaffold protein |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mohamed, A.; Taha, O.; El-Sherif, H.M.; Connerton, P.L.; Hooton, S.P.T.; Bassim, N.D.; Connerton, I.F.; El-Shibiny, A. Bacteriophage ZCSE2 is a Potent Antimicrobial against Salmonella enterica Serovars: Ultrastructure, Genomics and Efficacy. Viruses 2020, 12, 424. https://0-doi-org.brum.beds.ac.uk/10.3390/v12040424
Mohamed A, Taha O, El-Sherif HM, Connerton PL, Hooton SPT, Bassim ND, Connerton IF, El-Shibiny A. Bacteriophage ZCSE2 is a Potent Antimicrobial against Salmonella enterica Serovars: Ultrastructure, Genomics and Efficacy. Viruses. 2020; 12(4):424. https://0-doi-org.brum.beds.ac.uk/10.3390/v12040424
Chicago/Turabian StyleMohamed, Ahmed, Omar Taha, Hesham M. El-Sherif, Phillippa L. Connerton, Steven P.T. Hooton, Nabil D. Bassim, Ian F. Connerton, and Ayman El-Shibiny. 2020. "Bacteriophage ZCSE2 is a Potent Antimicrobial against Salmonella enterica Serovars: Ultrastructure, Genomics and Efficacy" Viruses 12, no. 4: 424. https://0-doi-org.brum.beds.ac.uk/10.3390/v12040424