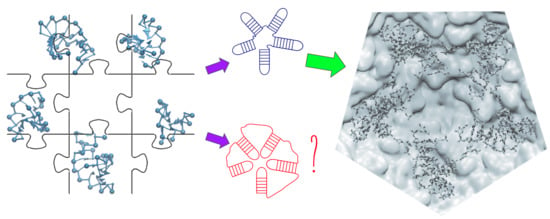
Structural 3D Domain Reconstruction of the RNA Genome from Viruses with Secondary Structure Models
Abstract
:1. Introduction
2. Materials and Methods
2.1. Coarse-Grained Model
2.2. Fragment Assembler
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hu, Y.; Zandi, R.; Anavitarte, A.; Knobler, C.M.; Gelbart, W.M. Packaging of a Polymer by a Viral Capsid: The Interplay Between Polymer Length and Capsid Size. Biophys. J. 2008, 94, 1428–1436. [Google Scholar] [CrossRef] [Green Version]
- Roos, W.H.; Gibbons, M.M.; Arkhipov, A.; Uetrecht, C.; Watts, N.R.; Wingfield, P.T.; Steven, A.C.; Heck, A.J.R.; Schulten, K.; Klug, W.S.; et al. Squeezing Protein Shells: How Continuum Elastic Models, Molecular Dynamics Simulations, and Experiments Coalesce at the Nanoscale. Biophys. J. 2010, 99, 1175–1181. [Google Scholar] [CrossRef] [Green Version]
- Castellanos, M.; Pérez, R.; Carrasco, C.; Hernando-Pérez, M.; Gómez-Herrero, J.; de Pablo, P.J.; Mateu, M.G. Mechanical Elasticity as a Physical Signature of Conformational Dynamics in a Virus Particle. Proc. Natl. Acad. Sci. USA 2012, 109, 12028–12033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stockley, P.G.; Ranson, N.A.; Twarock, R. A New Paradigm for the Roles of the Genome in ssRNA Viruses. Future Virol. 2013, 8, 531–543. [Google Scholar] [CrossRef]
- Garmann, R.F.; Comas-Garcia, M.; Koay, M.S.; Cornelissen, J.J.; Knobler, C.M.; Gelbart, W.M. Role of Electrostatics in the Assembly Pathway of a Single-Stranded RNA Virus. J. Virol. 2014, 88, 10472–10479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hernandez-Garcia, A.; Kraft, D.J.; Janssen, A.F.J.; Bomans, P.H.H.; Sommerdijk, N.A.J.M.; Thies-Weesie, D.M.E.; Favretto, M.E.; Brock, R.; de Wolf, F.A.; Werten, M.W.; et al. Design and Self-Assembly of Simple Coat Proteins for Artificial Viruses. Nat. Nanotechnol. 2014, 9, 698–702. [Google Scholar] [CrossRef]
- Perlmutter, J.D.; Hagan, M.F. The Role of Packaging Sites in Efficient and Specific Virus Assembly. J. Mol. Biol. 2015, 427, 2451–2467. [Google Scholar] [CrossRef] [Green Version]
- Beren, C.; Dreesens, L.L.; Liu, K.N.; Knobler, C.M.; Gelbart, W.M. The Effect of RNA Secondary Structure on the Self-Assembly of Viral Capsids. Biophys. J. 2017, 113, 339–347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Božič, A.L.; Micheletti, C.; Podgornik, R.; Tubiana, L. Compactness of Viral Genomes: Effect of Disperse and Localized Random Mutations. J. Phys. Condens. Matter 2018, 30, 084006. [Google Scholar] [CrossRef]
- Viso, J.F.; Belelli, P.; Machado, M.; González, H.; Pantano, S.; Amundarain, M.J.; Zamarreño, F.; Branda, M.M.; Guérin, D.M.; Costabel, M.D. Multiscale Modelization in a Small Virus: Mechanism of Proton Channeling and its Role in Triggering Capsid Disassembly. PLoS Comput. Biol. 2018, 14, e1006082. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martínez, M.; Cooper, C.D.; Poma, A.B.; Guzman, H.V. Free Energies of the Disassembly of Viral Capsids From a Multiscale Molecular Simulation Approach. J. Chem. Inf. Model. 2019, 60, 974–981. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Procyk, J.; Poppleton, E.; Šulc, P. Coarse-grained nucleic acid–protein model for hybrid nanotechnology. Soft Matter 2021, 17, 3586–3593. [Google Scholar] [CrossRef]
- Brion, P.; Westhof, E. Hierarchy and Dynamics of RNA Folding. Annu. Rev. Biophys. Biomol. Struct. 1997, 26, 113–137. [Google Scholar] [CrossRef]
- Tinoco, I.; Borer, P.N.; Dengler, B.; Levine, M.D.; Uhlenbeck, O.C.; Crothers, D.M.; Gralla, J. Improved Estimation of Secondary Structure in Ribonucleic Acids. Nature New Biol. 1973, 246, 40–41. [Google Scholar] [CrossRef]
- Tinoco, I.; Bustamante, C. How RNA Folds. J. Mol. Biol. 1999, 293, 271–281. [Google Scholar] [CrossRef]
- Mathews, D.H.; Sabina, J.; Zuker, M.; Turner, D.H. Expanded Sequence Dependence of Thermodynamic Parameters Improves Prediction of RNA Secondary Structure. J. Mol. Biol. 1999, 288, 911–940. [Google Scholar] [CrossRef] [Green Version]
- Bottaro, S.; Di Palma, F.; Bussi, G. The Role of Nucleobase Interactions in RNA Structure and Dynamics. Nucleic Acids Res. 2014, 42, 13306–13314. [Google Scholar] [CrossRef]
- Miao, Z.; Westhof, E. RNA Structure: Advances and Assessment of 3D Structure Prediction. Annu. Rev. Biophys. 2017, 46, 483–503. [Google Scholar] [CrossRef] [PubMed]
- Ponce-Salvatierra, A.; Astha; Merdas, K.; Nithin, C.; Ghosh, P.; Mukherjee, S.; Bujnicki, J.M. Computational modeling of RNA 3D structure based on experimental data. Biosci. Rep. 2019, 39, BSR20180430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, B.; Cao, Y.; Westhof, E.; Miao, Z. Advances in RNA 3D Structure Modeling Using Experimental Data. Front. Genet. 2020, 11, 1147. [Google Scholar] [CrossRef]
- Weeks, K.M. Advances in RNA structure analysis by chemical probing. Curr. Opin. Struct. Biol. 2010, 20, 295–304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mustoe, A.M.; Al-Hashimi, H.M.; Brooks, C.L. Secondary structure encodes a cooperative tertiary folding funnel in the Azoarcus ribozyme. Nucleic Acids Res. 2015, 44, 402–412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mustoe, A.M.; Lama, N.N.; Irving, P.S.; Olson, S.W.; Weeks, K.M. RNA base-pairing complexity in living cells visualized by correlated chemical probing. Proc. Natl. Acad. Sci. USA 2019, 116, 24574–24582. [Google Scholar] [CrossRef]
- Morandi, E.; Manfredonia, I.; Simon, L.M.; Anselmi, F.; van Hemert, M.J.; Oliviero, S.; Incarnato, D. Genome-scale deconvolution of RNA structure ensembles. Nat. Methods 2021, 18, 249–252. [Google Scholar] [CrossRef] [PubMed]
- Marinus, T.; Fessler, A.B.; Ogle, C.A.; Incarnato, D. A novel SHAPE reagent enables the analysis of RNA structure in living cells with unprecedented accuracy. Nucleic Acids Res. 2021, 49, e34. [Google Scholar] [CrossRef] [PubMed]
- Weidmann, C.A.; Mustoe, A.M.; Jariwala, P.B.; Calabrese, J.M.; Weeks, K.M. Analysis of RNA–protein networks with RNP-MaP defines functional hubs on RNA. Nat. Biotechnol. 2021, 39, 347–356. [Google Scholar] [CrossRef] [PubMed]
- Larman, B.C.; Dethoff, E.A.; Weeks, K.M. Packaged and Free Satellite Tobacco Mosaic Virus (STMV) RNA Genomes Adopt Distinct Conformational States. Biochemistry 2017, 56, 2175–2183. [Google Scholar] [CrossRef]
- Das, R.; Baker, D. Automated de novo prediction of native-like RNA tertiary structures. Proc. Natl. Acad. Sci. USA 2007, 104, 14664–14669. [Google Scholar] [CrossRef] [Green Version]
- Cheng, C.Y.; Chou, F.C.; Das, R. Chapter Two—Modeling Complex RNA Tertiary Folds with Rosetta. In Computational Methods for Understanding Riboswitches; Methods in Enzymology; Chen, S.J., Burke-Aguero, D.H., Eds.; Academic Press: Waltham, MA, USA, 2015; Volume 553, pp. 35–64. [Google Scholar] [CrossRef]
- Magnus, M.; Antczak, M.; Zok, T.; Wiedemann, J.; Lukasiak, P.; Cao, Y.; Bujnicki, J.M.; Westhof, E.; Szachniuk, M.; Miao, Z. RNA-Puzzles toolkit: A computational resource of RNA 3D structure benchmark datasets, structure manipulation, and evaluation tools. Nucleic Acids Res. 2019, 48, 576–588. [Google Scholar] [CrossRef]
- Schön, P. Atomic force microscopy of RNA: State of the art and recent advancements. Semin. Cell Dev. Biol. 2018, 73, 209–219. [Google Scholar] [CrossRef]
- Freddolino, P.L.; Arkhipov, A.S.; Larson, S.B.; McPherson, A.; Schulten, K. Molecular Dynamics Simulations of the Complete Satellite Tobacco Mosaic Virus. Structure 2006, 14, 437–449. [Google Scholar] [CrossRef] [Green Version]
- Zeng, Y.; Larson, S.B.; Heitsch, C.E.; McPherson, A.; Harvey, S.C. A Model for the Structure of Satellite Tobacco Mosaic Virus. J. Struct. Biol. 2012, 180, 110–116. [Google Scholar] [CrossRef] [Green Version]
- Cieplak, M.; Robbins, M.O. Nanoindentation of 35 Virus Capsids in a Molecular Model: Relating Mechanical Properties to Structure. PLoS ONE 2013, 8, e63640. [Google Scholar] [CrossRef] [Green Version]
- Schroeder, S.J. Probing Viral Genomic Structure: Alternative Viewpoints and Alternative Structures for Satellite Tobacco Mosaic Virus RNA. Biochemistry 2014, 53, 6728–6737. [Google Scholar] [CrossRef]
- Guzman, H.V.; Tretyakov, N.; Kobayashi, H.; Fogarty, A.C.; Kreis, K.; Krajniak, J.; Junghans, C.; Kremer, K.; Stuehn, T. ESPResSo++ 2.0: Advanced Methods for Multiscale Molecular Simulation. Comput. Phys. Commun. 2019, 238, 66–76. [Google Scholar] [CrossRef] [Green Version]
- Poblete, S.; Bottaro, S.; Bussi, G. A Nucleobase-Centered Coarse-Grained Representation for Structure Prediction of RNA Motifs. Nucleic Acids Res. 2017, 46, 1674–1683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poblete, S.; Bottaro, S.; Bussi, G. Effects and Limitations of a Nucleobase-Driven Backmapping Procedure for Nucleic Acids Using Steered Molecular Dynamics. Biochem. Biophys. Res. Commun. 2018, 498, 352–358. [Google Scholar] [CrossRef] [Green Version]
- Boniecki, M.J.; Lach, G.; Dawson, W.K.; Tomala, K.; Lukasz, P.; Soltysinski, T.; Rother, K.M.; Bujnicki, J.M. SimRNA: A Coarse-Grained Method for RNA Folding Simulations and 3D Structure Prediction. Nucleic Acids Res. 2015, 44, e63. [Google Scholar] [CrossRef] [PubMed]
- Šulc, P.; Romano, F.; Ouldridge, T.E.; Doye, J.P.K.; Louis, A.A. A Nucleotide-Level Coarse-Grained Model of RNA. J. Chem. Phys. 2014, 140, 235102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, D.; Chen, S.J. IsRNA: An Iterative Simulated Reference State Approach to Modeling Correlated Interactions in RNA Folding. J. Chem. Theory Comput. 2018, 14, 2230–2239. [Google Scholar] [CrossRef] [PubMed]
- Pasquali, S.; Derreumaux, P. HiRE-RNA: A High Resolution Coarse-Grained Energy Model for RNA. J. Phys. Chem. B 2010, 114, 11957–11966. [Google Scholar] [CrossRef]
- Kerpedjiev, P.; Siederdissen, C.H.Z.; Hofacker, I.L. Predicting RNA 3D structure using a coarse-grain helix-centered model. RNA 2015, 21, 1110–1121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perlmutter, J.D.; Hagan, M.F. Mechanisms of Virus Assembly. Annu. Rev. Phys. Chem. 2015, 66, 217–239. [Google Scholar] [CrossRef] [Green Version]
- Buzón, P.; Maity, S.; Roos, W.H. Physical virology: From virus self-assembly to particle mechanics. WIREs Nanomed. Nanobiotechnol. 2020, 12, e1613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schroeder, S.J. Perspectives on Viral RNA Genomes and the RNA Folding Problem. Viruses 2020, 12, 1126. [Google Scholar] [CrossRef]
- Zandi, R.; Dragnea, B.; Travesset, A.; Podgornik, R. On Virus Growth and Form. Phys. Rep. 2020, 847, 1–102. [Google Scholar] [CrossRef]
- Dai, X.; Li, Z.; Lai, M.; Shu, S.; Du, Y.; Zhou, Z.H.; Sun, R. In situ structures of the genome and genome-delivery apparatus in a single-stranded RNA virus. Nature 2017, 541, 112–116. [Google Scholar] [CrossRef] [Green Version]
- Beren, C.; Cui, Y.; Chakravarty, A.; Yang, X.; Rao, A.L.N.; Knobler, C.M.; Zhou, Z.H.; Gelbart, W.M. Genome organization and interaction with capsid protein in a multipartite RNA virus. Proc. Natl. Acad. Sci. USA 2020, 117, 10673–10680. [Google Scholar] [CrossRef]
- Tomezsko, P.J.; Corbin, V.D.A.; Gupta, P.; Swaminathan, H.; Glasgow, M.; Persad, S.; Edwards, M.D.; Mcintosh, L.; Papenfuss, A.T.; Emery, A.; et al. Determination of RNA structural diversity and its role in HIV-1 RNA splicing. Nature 2020, 582, 438–442. [Google Scholar] [CrossRef]
- Chojnowski, G.; Zaborowski, R.; Magnus, M.; Bujnicki, J.M. RNA fragment assembly with experimental restraints. bioRxiv 2021. [Google Scholar] [CrossRef]
- Rangan, R.; Watkins, A.M.; Chacon, J.; Kretsch, R.; Kladwang, W.; Zheludev, I.N.; Townley, J.; Rynge, M.; Thain, G.; Das, R. De novo 3D models of SARS-CoV-2 RNA elements from consensus experimental secondary structures. Nucleic Acids Res. 2021, 49, 3092–3108. [Google Scholar] [CrossRef] [PubMed]
- Kappel, K.; Zhang, K.; Su, Z.; Watkins, A.M.; Kladwang, W.; Li, S.; Pintilie, G.; Topkar, V.V.; Rangan, R.; Zheludev, I.N.; et al. Accelerated cryo-EM-guided determination of three-dimensional RNA-only structures. Nat. Methods 2020, 17, 699–707. [Google Scholar] [CrossRef] [PubMed]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Poblete, S.; Guzman, H.V. Structural 3D Domain Reconstruction of the RNA Genome from Viruses with Secondary Structure Models. Viruses 2021, 13, 1555. https://0-doi-org.brum.beds.ac.uk/10.3390/v13081555
Poblete S, Guzman HV. Structural 3D Domain Reconstruction of the RNA Genome from Viruses with Secondary Structure Models. Viruses. 2021; 13(8):1555. https://0-doi-org.brum.beds.ac.uk/10.3390/v13081555
Chicago/Turabian StylePoblete, Simón, and Horacio V. Guzman. 2021. "Structural 3D Domain Reconstruction of the RNA Genome from Viruses with Secondary Structure Models" Viruses 13, no. 8: 1555. https://0-doi-org.brum.beds.ac.uk/10.3390/v13081555