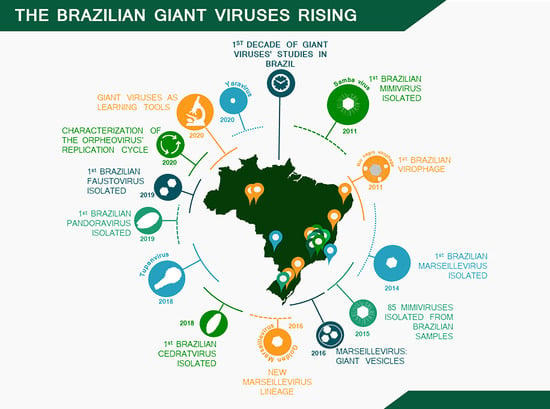
A Brief History of Giant Viruses’ Studies in Brazilian Biomes
Abstract
:1. Introduction
2. Giant Viruses Discovery and Isolation
2.1. Mimiviruses Boosted Amoebal Giant Viruses’ Research
2.2. The Second Family Arises: The Discovery of Marseilleviruses
2.3. Opening the GVs’ Box: The Discovery of Pandoraviruses
2.4. A Double-Corked GV: Isolation and Characterization of the Cedratviruses
2.5. Another Amoeba, Another Virus: Discovery and Characterization of Orpheovirus
2.6. The Isolation and Characterization of Faustoviruses
2.7. Yaravirus, a Small Virus among the Giants
3. A Fight for Supremacy: Peculiar Features of GVs and Their Interaction with Amoeba Hosts
4. Giant Viruses As a Tool to Update and Inspire: From the Research Fields to the Classroom
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Scola, B.L.; Audic, S.; Robert, C.; Jungang, L.; de Lamballerie, X.; Drancourt, M.; Birtles, R.; Claverie, J.-M.; Raoult, D. A Giant Virus in Amoebae. Science 2003, 299, 2033. [Google Scholar] [CrossRef]
- Iyer, L.M.; Aravind, L.; Koonin, E.V. Common Origin of Four Diverse Families of Large Eukaryotic DNA Viruses. J. Virol. 2001, 75, 11720–11734. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodrigues, R.A.; Andrade, A.C.; Boratto, P.V.d.M.; Trindade, G.d.S.; Kroon, E.G.; Abrahão, J.S. An Anthropocentric View of the Virosphere-Host Relationship. Front. Microbiol. 2017, 8, 1673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- La Scola, B.; Desnues, C.; Pagnier, I.; Robert, C.; Barrassi, L.; Fournous, G.; Merchat, M.; Suzan-Monti, M.; Forterre, P.; Koonin, E.; et al. The Virophage as a Unique Parasite of the Giant Mimivirus. Nature 2008, 455, 100–104. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V.; Yutin, N. Origin and Evolution of Eukaryotic Large Nucleo-Cytoplasmic DNA Viruses. Intervirology 2010, 53, 284–292. [Google Scholar] [CrossRef] [Green Version]
- Boyer, M.; Yutin, N.; Pagnier, I.; Barrassi, L.; Fournous, G.; Espinosa, L.; Robert, C.; Azza, S.; Sun, S.; Rossmann, M.G.; et al. Giant Marseillevirus Highlights the Role of Amoebae as a Melting Pot in Emergence of Chimeric Microorganisms. Proc. Natl. Acad. Sci. USA 2009, 106, 21848–21853. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andrade, A.C.D.S.P.; Arantes, T.S.; Rodrigues, R.A.L.; Machado, T.B.; Dornas, F.P.; Landell, M.F.; Furst, C.; Borges, L.G.A.; Dutra, L.A.L.; Almeida, G.; et al. Ubiquitous Giants: A Plethora of Giant Viruses Found in Brazil and Antarctica. Virol. J. 2018, 15, 22. [Google Scholar] [CrossRef] [Green Version]
- Aherfi, S.; Colson, P.; La Scola, B.; Raoult, D. Giant Viruses of Amoebas: An Update. Front. Microbiol. 2016, 7, 349. [Google Scholar] [CrossRef] [Green Version]
- Schulz, F.; Yutin, N.; Ivanova, N.N.; Ortega, D.R.; Lee, T.K.; Vierheilig, J.; Daims, H.; Horn, M.; Wagner, M.; Jensen, G.J.; et al. Giant Viruses with an Expanded Complement of Translation System Components. Science 2017, 356, 82–85. [Google Scholar] [CrossRef] [Green Version]
- Abrahão, J.; Silva, L.; Silva, L.S.; Khalil, J.Y.B.; Rodrigues, R.; Arantes, T.; Assis, F.; Boratto, P.; Andrade, M.; Kroon, E.G.; et al. Tailed Giant Tupanvirus Possesses the Most Complete Translational Apparatus of the Known Virosphere. Nat. Commun. 2018, 9, 749. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshikawa, G.; Blanc-Mathieu, R.; Song, C.; Kayama, Y.; Mochizuki, T.; Murata, K.; Ogata, H.; Takemura, M. Medusavirus, a Novel Large DNA Virus Discovered from Hot Spring Water. J. Virol. 2019, 93, e02130-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Legendre, M.; Lartigue, A.; Bertaux, L.; Jeudy, S.; Bartoli, J.; Lescot, M.; Alempic, J.-M.; Ramus, C.; Bruley, C.; Labadie, K.; et al. In-Depth Study of Mollivirus Sibericum, a New 30,000-y-Old Giant Virus Infecting Acanthamoeba. Proc. Natl. Acad. Sci. USA 2015, 112, E5327–E5335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abrahão, J.S.; Dornas, F.P.; Silva, L.C.; Almeida, G.M.; Boratto, P.V.; Colson, P.; La Scola, B.; Kroon, E.G. Acanthamoeba Polyphaga Mimivirus and Other Giant Viruses: An Open Field to Outstanding Discoveries. Virol. J. 2014, 11, 120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marciano-Cabral, F.; Cabral, G. Acanthamoeba spp. as Agents of Disease in Humans. Clin. Microbiol. Rev. 2003, 16, 273–307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colson, P.; La Scola, B.; Raoult, D. Giant Viruses of Amoebae: A Journey through Innovative Research and Paradigm Changes. Annu. Rev. Virol. 2017, 4, 61–85. [Google Scholar] [CrossRef]
- Andrade, K.R.; Boratto, P.P.V.M.; Rodrigues, F.P.; Silva, L.C.F.; Dornas, F.P.; Pilotto, M.R.; La Scola, B.; Almeida, G.M.F.; Kroon, E.G.; Abrahão, J.S. Oysters as Hot Spots for Mimivirus Isolation. Arch. Virol. 2015, 160, 477–482. [Google Scholar] [CrossRef]
- Mihara, T.; Koyano, H.; Hingamp, P.; Grimsley, N.; Goto, S.; Ogata, H. Taxon Richness of “Megaviridae” Exceeds Those of Bacteria and Archaea in the Ocean. Microbes Environ. 2018, 33, 162–171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moniruzzaman, M.; Martinez-Gutierrez, C.A.; Weinheimer, A.R.; Aylward, F.O. Dynamic Genome Evolution and Complex Virocell Metabolism of Globally-Distributed Giant Viruses. Nat. Commun. 2020, 11, 1710. [Google Scholar] [CrossRef] [Green Version]
- Moniruzzaman, M.; Weinheimer, A.R.; Martinez-Gutierrez, C.A.; Aylward, F.O. Widespread Endogenization of Giant Viruses Shapes Genomes of Green Algae. Nature 2020, 588, 141–145. [Google Scholar] [CrossRef]
- Marcelino, V.M.; Espinola, M.V.P.C.; Serrano-Solis, V.; Farias, S.T. Evolution of the Genus Mimivirus Based on Translation Protein Homology and Its Implication in the Tree of Life. Genet. Mol. Res. 2017, 16, 1–7. [Google Scholar] [CrossRef]
- Dornas, F.P.; Khalil, J.Y.B.; Pagnier, I.; Raoult, D.; Abrahão, J.; La Scola, B. Isolation of New Brazilian Giant Viruses from Environmental Samples Using a Panel of Protozoa. Front. Microbiol. 2015, 6, 1086. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campos, R.K.; Boratto, P.V.; Assis, F.L.; Aguiar, E.R.; Silva, L.C.; Albarnaz, J.D.; Dornas, F.P.; Trindade, G.S.; Ferreira, P.P.; Marques, J.T.; et al. Samba Virus: A Novel Mimivirus from a Giant Rain Forest, the Brazilian Amazon. Virol. J. 2014, 11, 95. [Google Scholar] [CrossRef] [Green Version]
- Guerreiro, R.L.; Bergier, I.; McGlue, M.M.; Warren, L.V.; de Abreu, U.G.P.; Abrahão, J.; Assine, M.L. The Soda Lakes of Nhecolândia: A Conservation Opportunity for the Pantanal Wetlands. Perspect. Ecol. Conserv. 2019, 17, 9–18. [Google Scholar] [CrossRef]
- Dos Santos Silva, L.K.; Andrade, A.C.S.P.; Dornas, F.P.; Rodrigues, R.A.L.; Arantes, T.; Kroon, E.G.; Bonjardim, C.A.; Abrahão, J.S. Cedratvirus Getuliensis Replication Cycle: An in-Depth Morphological Analysis. Sci. Rep. 2018, 8, 4000. [Google Scholar] [CrossRef]
- Boratto, P.V.M.; Arantes, T.S.; Silva, L.C.F.; Assis, F.L.; Kroon, E.G.; La Scola, B.; Abrahão, J.S. Niemeyer Virus: A New Mimivirus Group A Isolate Harboring a Set of Duplicated Aminoacyl-TRNA Synthetase Genes. Front. Microbiol. 2015, 6, 1256. [Google Scholar] [CrossRef] [Green Version]
- Borges, I.; Rodrigues, R.A.L.; Dornas, F.P.; Almeida, G.; Aquino, I.; Bonjardim, C.A.; Kroon, E.G.; La Scola, B.; Abrahão, J.S. Trapping the Enemy: Vermamoeba vermiformis Circumvents Faustovirus Mariensis Dissemination by Enclosing Viral Progeny inside Cysts. J. Virol. 2019, 93, e00312-19. [Google Scholar] [CrossRef] [Green Version]
- Boratto, P.V.M.; Oliveira, G.P.; Machado, T.B.; Andrade, A.C.S.P.; Baudoin, J.-P.; Klose, T.; Schulz, F.; Azza, S.; Decloquement, P.; Chabrière, E.; et al. Yaravirus: A Novel 80-Nm Virus Infecting Acanthamoeba Castellanii. Proc. Natl. Acad. Sci. USA 2020, 117, 16579–16586. [Google Scholar] [CrossRef]
- Schrad, J.R.; Young, E.J.; Abrahão, J.S.; Cortines, J.R.; Parent, K.N. Microscopic Characterization of the Brazilian Giant Samba Virus. Viruses 2017, 9, 30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borges, I.A.; de Assis, F.L.; Silva, L.K.D.S.; Abrahão, J. Rio Negro Virophage: Sequencing of the near Complete Genome and Transmission Electron Microscopy of Viral Factories and Particles. Braz. J. Microbiol. 2018, 49 (Suppl. S1), 260–261. [Google Scholar] [CrossRef] [PubMed]
- Mougari, S.; Bekliz, M.; Abrahao, J.; Di Pinto, F.; Levasseur, A.; La Scola, B. Guarani Virophage, a New Sputnik-Like Isolate From a Brazilian Lake. Front. Microbiol. 2019, 10, 1003. [Google Scholar] [CrossRef] [Green Version]
- Assis, F.L.; Franco-Luiz, A.P.M.; Dos Santos, R.N.; Campos, F.S.; Dornas, F.P.; Boratto, P.V.M.; Franco, A.C.; Abrahao, J.S.; Colson, P.; Scola, B.L. Genome Characterization of the First Mimiviruses of Lineage C Isolated in Brazil. Front. Microbiol. 2017, 8, 2562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boratto, P.V.M.; Dornas, F.P.; da Silva, L.C.F.; Rodrigues, R.A.L.; Oliveira, G.P.; Cortines, J.R.; Drumond, B.P.; Abrahão, J.S. Analyses of the Kroon Virus Major Capsid Gene and Its Transcript Highlight a Distinct Pattern of Gene Evolution and Splicing among Mimiviruses. J. Virol. 2018, 92, e01782-17. [Google Scholar] [CrossRef] [Green Version]
- Assis, F.L.; Bajrai, L.; Abrahao, J.S.; Kroon, E.G.; Dornas, F.P.; Andrade, K.R.; Boratto, P.V.M.; Pilotto, M.R.; Robert, C.; Benamar, S.; et al. Pan-Genome Analysis of Brazilian Lineage A Amoebal Mimiviruses. Viruses 2015, 7, 3483–3499. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodrigues, R.A.L.; Mougari, S.; Colson, P.; La Scola, B.; Abrahão, J.S. “Tupanvirus”, a New Genus in the Family Mimiviridae. Arch. Virol. 2019, 164, 325–331. [Google Scholar] [CrossRef] [Green Version]
- De Miranda Boratto, P.V.; Dos Santos Pereira Andrade, A.C.; Araújo Lima Rodrigues, R.; La Scola, B.; Santos Abrahão, J. The Multiple Origins of Proteins Present in Tupanvirus Particles. Curr. Opin. Virol. 2019, 36, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Raoult, D. The Post-Darwinist Rhizome of Life. Lancet 2010, 375, 104–105. [Google Scholar] [CrossRef]
- Abrahão, J.S.; Araújo, R.; Colson, P.; Scola, B.L. The Analysis of Translation-Related Gene Set Boosts Debates around Origin and Evolution of Mimiviruses. PLoS Genet. 2017, 13, e1006532. [Google Scholar] [CrossRef]
- Aherfi, S.; La Scola, B.; Pagnier, I.; Raoult, D.; Colson, P. The Expanding Family Marseilleviridae. Virology 2014, 466–467, 27–37. [Google Scholar] [CrossRef] [Green Version]
- Thomas, V.; Bertelli, C.; Collyn, F.; Casson, N.; Telenti, A.; Goesmann, A.; Croxatto, A.; Greub, G. Lausannevirus, a Giant Amoebal Virus Encoding Histone Doublets. Environ. Microbiol. 2011, 13, 1454–1466. [Google Scholar] [CrossRef] [Green Version]
- Aherfi, S.; Pagnier, I.; Fournous, G.; Raoult, D.; La Scola, B.; Colson, P. Complete Genome Sequence of Cannes 8 Virus, a New Member of the Proposed Family “Marseilleviridae”. Virus Genes 2013, 47, 550–555. [Google Scholar] [CrossRef]
- Pagnier, I.; Reteno, D.-G.I.; Saadi, H.; Boughalmi, M.; Gaia, M.; Slimani, M.; Ngounga, T.; Bekliz, M.; Colson, P.; Raoult, D.; et al. A Decade of Improvements in Mimiviridae and Marseilleviridae Isolation from Amoeba. Intervirology 2013, 56, 354–363. [Google Scholar] [CrossRef]
- Aherfi, S.; Boughalmi, M.; Pagnier, I.; Fournous, G.; La Scola, B.; Raoult, D.; Colson, P. Complete Genome Sequence of Tunisvirus, a New Member of the Proposed Family Marseilleviridae. Arch. Virol. 2014, 159, 2349–2358. [Google Scholar] [CrossRef] [PubMed]
- Boughalmi, M.; Pagnier, I.; Aherfi, S.; Colson, P.; Raoult, D.; La Scola, B. First Isolation of a Marseillevirus in the Diptera Syrphidae Eristalis Tenax. Intervirology 2013, 56, 386–394. [Google Scholar] [CrossRef]
- Lagier, J.-C.; Armougom, F.; Million, M.; Hugon, P.; Pagnier, I.; Robert, C.; Bittar, F.; Fournous, G.; Gimenez, G.; Maraninchi, M.; et al. Microbial Culturomics: Paradigm Shift in the Human Gut Microbiome Study. Clin. Microbiol. Infect. 2012, 18, 1185–1193. [Google Scholar] [CrossRef] [Green Version]
- Colson, P.; Fancello, L.; Gimenez, G.; Armougom, F.; Desnues, C.; Fournous, G.; Yoosuf, N.; Million, M.; La Scola, B.; Raoult, D. Evidence of the Megavirome in Humans. J. Clin. Virol. 2013, 57, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Doutre, G.; Philippe, N.; Abergel, C.; Claverie, J.-M. Genome Analysis of the First Marseilleviridae Representative from Australia Indicates That Most of Its Genes Contribute to Virus Fitness. J. Virol. 2014, 88, 14340–14349. [Google Scholar] [CrossRef] [Green Version]
- Dornas, F.P.; Assis, F.L.; Aherfi, S.; Arantes, T.; Abrahão, J.S.; Colson, P.; La Scola, B. A Brazilian Marseillevirus Is the Founding Member of a Lineage in Family Marseilleviridae. Viruses 2016, 8, 76. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edgar, R.C. MUSCLE: A Multiple Sequence Alignment Method with Reduced Time and Space Complexity. BMC Bioinform. 2004, 5, 113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. TrimAl: A Tool for Automated Alignment Trimming in Large-Scale Phylogenetic Analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (ITOL) v5: An Online Tool for Phylogenetic Tree Display and Annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Dos Santos, R.N.; Campos, F.S.; Medeiros de Albuquerque, N.R.; Finoketti, F.; Côrrea, R.A.; Cano-Ortiz, L.; Assis, F.L.; Arantes, T.S.; Roehe, P.M.; Franco, A.C. A New Marseillevirus Isolated in Southern Brazil from Limnoperna Fortunei. Sci. Rep. 2016, 6, 35237. [Google Scholar] [CrossRef] [Green Version]
- Philippe, N.; Legendre, M.; Doutre, G.; Couté, Y.; Poirot, O.; Lescot, M.; Arslan, D.; Seltzer, V.; Bertaux, L.; Bruley, C.; et al. Pandoraviruses: Amoeba Viruses with Genomes up to 2.5 Mb Reaching That of Parasitic Eukaryotes. Science 2013, 341, 281–286. [Google Scholar] [CrossRef] [Green Version]
- Scheid, P.; Balczun, C.; Schaub, G.A. Some Secrets Are Revealed: Parasitic Keratitis Amoebae as Vectors of the Scarcely Described Pandoraviruses to Humans. Parasitol. Res. 2014, 113, 3759–3764. [Google Scholar] [CrossRef] [PubMed]
- Antwerpen, M.H.; Georgi, E.; Zoeller, L.; Woelfel, R.; Stoecker, K.; Scheid, P. Whole-Genome Sequencing of a Pandoravirus Isolated from Keratitis-Inducing Acanthamoeba. Genome Announc. 2015, 3, e00136-15. [Google Scholar] [CrossRef] [Green Version]
- Legendre, M.; Alempic, J.-M.; Philippe, N.; Lartigue, A.; Jeudy, S.; Poirot, O.; Ta, N.T.; Nin, S.; Couté, Y.; Abergel, C.; et al. Pandoravirus Celtis Illustrates the Microevolution Processes at Work in the Giant Pandoraviridae Genomes. Front. Microbiol. 2019, 10, 430. [Google Scholar] [CrossRef] [Green Version]
- Legendre, M.; Fabre, E.; Poirot, O.; Jeudy, S.; Lartigue, A.; Alempic, J.-M.; Beucher, L.; Philippe, N.; Bertaux, L.; Christo-Foroux, E.; et al. Diversity and Evolution of the Emerging Pandoraviridae Family. Nat. Commun. 2018, 9, 2285. [Google Scholar] [CrossRef]
- Pereira Andrade, A.C.D.S.; Victor de Miranda Boratto, P.; Rodrigues, R.A.L.; Bastos, T.M.; Azevedo, B.L.; Dornas, F.P.; Oliveira, D.B.; Drumond, B.P.; Kroon, E.G.; Abrahão, J.S. New Isolates of Pandoraviruses: Contribution to the Study of Replication Cycle Steps. J. Virol. 2019, 93, e01942-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andreani, J.; Aherfi, S.; Bou Khalil, J.Y.; Di Pinto, F.; Bitam, I.; Raoult, D.; Colson, P.; La Scola, B. Cedratvirus, a Double-Cork Structured Giant Virus, Is a Distant Relative of Pithoviruses. Viruses 2016, 8, 300. [Google Scholar] [CrossRef] [Green Version]
- Bertelli, C.; Mueller, L.; Thomas, V.; Pillonel, T.; Jacquier, N.; Greub, G. Cedratvirus Lausannensis—Digging into Pithoviridae Diversity. Environ. Microbiol. 2017, 19, 4022–4034. [Google Scholar] [CrossRef] [Green Version]
- Jeudy, S.; Rigou, S.; Alempic, J.-M.; Claverie, J.-M.; Abergel, C.; Legendre, M. The DNA Methylation Landscape of Giant Viruses. Nat. Commun. 2020, 11, 2657. [Google Scholar] [CrossRef]
- Andreani, J.; Khalil, J.Y.B.; Baptiste, E.; Hasni, I.; Michelle, C.; Raoult, D.; Levasseur, A.; La Scola, B. Orpheovirus IHUMI-LCC2: A New Virus among the Giant Viruses. Front. Microbiol. 2018, 8, 2643. [Google Scholar] [CrossRef] [Green Version]
- Rodrigues, R.A.L.; Andreani, J.; Andrade, A.C.D.S.P.; Machado, T.B.; Abdi, S.; Levasseur, A.; Abrahão, J.S.; La Scola, B. Morphologic and Genomic Analyses of New Isolates Reveal a Second Lineage of Cedratviruses. J. Virol. 2018, 92, e00372-18. [Google Scholar] [CrossRef] [Green Version]
- Souza, F.; Rodrigues, R.; Reis, E.; Lima, M.; La Scola, B.; Abrahão, J. In-Depth Analysis of the Replication Cycle of Orpheovirus. Virol. J. 2019, 16, 158. [Google Scholar] [CrossRef]
- Reteno, D.G.; Benamar, S.; Khalil, J.B.; Andreani, J.; Armstrong, N.; Klose, T.; Rossmann, M.; Colson, P.; Raoult, D.; La Scola, B. Faustovirus, an Asfarvirus-Related New Lineage of Giant Viruses Infecting Amoebae. J. Virol. 2015, 89, 6585–6594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oliveira, G.P.; de Aquino, I.L.M.; Luiz, A.P.M.F.; Abrahão, J.S. Putative Promoter Motif Analyses Reinforce the Evolutionary Relationships Among Faustoviruses, Kaumoebavirus, and Asfarvirus. Front. Microbiol. 2018, 9, 1041. [Google Scholar] [CrossRef] [PubMed]
- Tully, B.J.; Graham, E.D.; Heidelberg, J.F. The Reconstruction of 2,631 Draft Metagenome-Assembled Genomes from the Global Oceans. Sci. Data 2018, 5, 170203. [Google Scholar] [CrossRef] [Green Version]
- Schulz, F.; Roux, S.; Paez-Espino, D.; Jungbluth, S.; Walsh, D.A.; Denef, V.J.; McMahon, K.D.; Konstantinidis, K.T.; Eloe-Fadrosh, E.A.; Kyrpides, N.C.; et al. Giant Virus Diversity and Host Interactions through Global Metagenomics. Nature 2020, 578, 432–436. [Google Scholar] [CrossRef]
- Hingamp, P.; Grimsley, N.; Acinas, S.G.; Clerissi, C.; Subirana, L.; Poulain, J.; Ferrera, I.; Sarmento, H.; Villar, E.; Lima-Mendez, G.; et al. Exploring Nucleo-Cytoplasmic Large DNA Viruses in Tara Oceans Microbial Metagenomes. ISME J. 2013, 7, 1678–1695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodrigues, R.A.L.; dos Santos Silva, L.K.; Dornas, F.P.; de Oliveira, D.B.; Magalhães, T.F.F.; Santos, D.A.; Costa, A.O.; de Macêdo Farias, L.; Magalhães, P.P.; Bonjardim, C.A.; et al. Mimivirus Fibrils Are Important for Viral Attachment to the Microbial World by a Diverse Glycoside Interaction Repertoire. J. Virol. 2015, 89, 11812–11819. [Google Scholar] [CrossRef] [Green Version]
- Ghigo, E.; Kartenbeck, J.; Lien, P.; Pelkmans, L.; Capo, C.; Mege, J.-L.; Raoult, D. Ameobal Pathogen Mimivirus Infects Macrophages through Phagocytosis. PLoS Pathog. 2008, 4, e1000087. [Google Scholar] [CrossRef] [Green Version]
- Andrade, A.C.D.S.P.; Rodrigues, R.A.L.; Oliveira, G.P.; Andrade, K.R.; Bonjardim, C.A.; La Scola, B.; Kroon, E.G.; Abrahão, J.S. Filling Knowledge Gaps for Mimivirus Entry, Uncoating, and Morphogenesis. J. Virol. 2017, 91, e01335-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schrad, J.R.; Abrahão, J.S.; Cortines, J.R.; Parent, K.N. Structural and Proteomic Characterization of the Initiation of Giant Virus Infection. Cell 2020, 181, 1046–1061.e6. [Google Scholar] [CrossRef] [PubMed]
- De Souza, G.A.P.; Queiroz, V.F.; Coelho, L.F.L.; Abrahão, J.S. Alohomora! What the Entry Mechanisms Tell Us about the Evolution and Diversification of Giant Viruses and Their Hosts. Curr. Opin. Virol. 2021, 47, 79–85. [Google Scholar] [CrossRef] [PubMed]
- Quemin, E.R.; Corroyer-Dulmont, S.; Krijnse-Locker, J. Entry and Disassembly of Large DNA Viruses: Electron Microscopy Leads the Way. J. Mol. Biol. 2018, 430, 1714–1724. [Google Scholar] [CrossRef]
- Parent, K.N.; Schrad, J.R.; Young, E.J.; Abrahao, J.S.; Cortines, J.R. A Gateway into Understanding the Unique Vertex of Samba Virus. Microsc. Microanal. 2018, 24, 1438–1439. [Google Scholar] [CrossRef] [Green Version]
- Zauberman, N.; Mutsafi, Y.; Halevy, D.B.; Shimoni, E.; Klein, E.; Xiao, C.; Sun, S.; Minsky, A. Distinct DNA Exit and Packaging Portals in the Virus Acanthamoeba Polyphaga Mimivirus. PLoS Biol. 2008, 6, e114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oliveira, G.; Silva, L.; Leão, T.; Mougari, S.; da Fonseca, F.G.; Kroon, E.G.; La Scola, B.; Abrahão, J.S. Tupanvirus-Infected Amoebas Are Induced to Aggregate with Uninfected Cells Promoting Viral Dissemination. Sci. Rep. 2019, 9, 183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Farias, S.T.; Jheeta, S.; Prosdocimi, F. Viruses as a Survival Strategy in the Armory of Life. Hist. Philos. Life Sci. 2019, 41, 45. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, G.; La Scola, B.; Abrahão, J. Giant Virus vs Amoeba: Fight for Supremacy. Virol. J. 2019, 16, 126. [Google Scholar] [CrossRef] [Green Version]
- Boratto, P.; Albarnaz, J.D.; Almeida, G.M.; Botelho, L.; Fontes, A.C.L.; Costa, A.O.; Santos, D.d.A.; Bonjardim, C.A.; La Scola, B.; Kroon, E.G.; et al. Acanthamoeba Polyphaga Mimivirus Prevents Amoebal Encystment-Mediating Serine Proteinase Expression and Circumvents Cell Encystment. J. Virol. 2015, 89, 2962–2965. [Google Scholar] [CrossRef] [Green Version]
- Silva, L.K.D.S.; Boratto, P.V.M.; La Scola, B.; Bonjardim, C.A.; Abrahão, J.S. Acanthamoeba and Mimivirus Interactions: The Role of Amoebal Encystment and the Expansion of the “Cheshire Cat” Theory. Curr. Opin. Microbiol. 2016, 31, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Korn, E.D.; Weisman, R.A. Phagocytosis of Latex Beads by Acanthamoeba. II. Electron Microscopic Study of the Initial Events. J. Cell Biol. 1967, 34, 219–227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arantes, T.S.; Rodrigues, R.A.L.; Dos Santos Silva, L.K.; Oliveira, G.P.; de Souza, H.L.; Khalil, J.Y.B.; de Oliveira, D.B.; Torres, A.A.; da Silva, L.L.; Colson, P.; et al. The Large Marseillevirus Explores Different Entry Pathways by Forming Giant Infectious Vesicles. J. Virol. 2016, 90, 5246–5255. [Google Scholar] [CrossRef] [Green Version]
- Oliveira, G.P.; Lima, M.T.; Arantes, T.S.; Assis, F.L.; Rodrigues, R.A.L.; da Fonseca, F.G.; Bonjardim, C.A.; Kroon, E.G.; Colson, P.; La Scola, B.; et al. The Investigation of Promoter Sequences of Marseilleviruses Highlights a Remarkable Abundance of the AAATATTT Motif in Intergenic Regions. J. Virol. 2017, 91, e01088-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodrigues, R.A.L.; Louazani, A.C.; Picorelli, A.; Oliveira, G.P.; Lobo, F.P.; Colson, P.; La Scola, B.; Abrahão, J.S. Analysis of a Marseillevirus Transcriptome Reveals Temporal Gene Expression Profile and Host Transcriptional Shift. Front. Microbiol. 2020, 11, 651. [Google Scholar] [CrossRef] [Green Version]
- Serrano-Solís, V.; Toscano Soares, P.E.; de Farías, S.T. Genomic Signatures Among Acanthamoeba Polyphaga Entoorganisms Unveil Evidence of Coevolution. J. Mol. Evol. 2019, 87, 7–15. [Google Scholar] [CrossRef]
- Akashi, M.; Fukaya, S.; Uchiyama, C.; Aoki, K.; Takemura, M. Visualization of Giant Virus Particles and Development of “VIRAMOS” for High School and University Biology Course. Biochem. Mol. Biol. Educ. 2019, 47, 426–431. [Google Scholar] [CrossRef]
- Paez-Espino, D.; Eloe-Fadrosh, E.A.; Pavlopoulos, G.A.; Thomas, A.D.; Huntemann, M.; Mikhailova, N.; Rubin, E.; Ivanova, N.N.; Kyrpides, N.C. Uncovering Earth’s Virome. Nature 2016, 536, 425–430. [Google Scholar] [CrossRef]
- Matza-Porges, S.; Nathan, D. A Biosafety Level 2 Virology Lab for Biotechnology Undergraduates. Biochem. Mol. Biol. Educ. 2017, 45, 537–543. [Google Scholar] [CrossRef] [Green Version]
- De Souza, G.A.P.; Queiroz, V.F.; Lima, M.T.; de Sousa Reis, E.V.; Coelho, L.F.L.; Abrahão, J.S. Virus Goes Viral: An Educational Kit for Virology Classes. Virol. J. 2020, 17, 13. [Google Scholar] [CrossRef] [PubMed]






| Group of Virus | Virus | Type of Sample | Location (Year of Isolation) | Genome Size (bp) | ORFs | ORFans | GC % | Reference |
|---|---|---|---|---|---|---|---|---|
| Samba virus | Fresh water | Negro River (2011) | 1,181,380 | 971 | 0 | 27 | Campos et al., 2014 | |
| Amazonia virus | Fresh water | Negro River (2011) | 1,179,119 | 979 | 1 (0.1%) | 27 | Assis et al., 2015 | |
| Mimiviridae | Kroon virus | Urban lake water | Lagoa Santa city (2012) | 1,221,932 | 944 | 3 (0.3%) | 27 | Assis et al., 2015 |
| (lineage A mimivirus) | Oyster virus | Oysters | Santa Catarina state (2013) | 1,200,220 | 948 | 1 (0.1%) | 27 | Assis et al., 2015 |
| Niemeyer virus | Urban lake water | Pampulha Lagoon (2011) | 1,299,140 | 1003 | 0 | 28 | Boratto et al., 2015 | |
| Mimiviridae | Borely moumouvirus | Fresh Water | Serra do Cipó (2018) | 1,038,187 | 947 | 3 (0.3%) | 25.2 | Silva et al., 2020 |
| (lineage B mimivirus) | ||||||||
| Mimiviridae | Mimivirus gilmour | Urban lake water | Pampulha Lagoon (2014) | 1,258,663 | 1135 | 28 (2.4%) | 26 | Assis et al., 2017 |
| (lineage C mimivirus) | Mimivirus golden | Golden mussels | Guaíba Lake (2014) | 1,248,960 | 1127 | 19 (1.6%) | 26 | Assis et al., 2017 |
| Mimiviridae | Tupanvirus deep ocean | Deep Ocean sediments | Campos dos Goytacazes city (2018) | 1,439,508 | 1276 | 378 (29.6%) | 28 | Abrahão et al., 2018 |
| Tupanvirus soda lake | Soda Lake | Nhecolândia, Pantanal biome (2018) | 1,516,267 | 1359 | 375 (27.6%) | 28 | Abrahão et al., 2018 | |
| Marseilleviridae | Brazilian marseillevirus | Sewage | Pampulha Lagoon (2014) | 362,276 | 491 | 29 (5.9%) | 43.3 | Dornas et al., 2016 |
| Golden marseillevirus | Golden mussels | Guaíba Lake (2014) | 360,610 | 483 | 43 (8.9%) | 43.1 | Santos et al., 2016 | |
| Cedratviruses | Brazilian cedratvirus | Water supplemented with biofloc | Belo Horizonte city (2018) | 460,038 | 533 | 11 (2.1%) | 42.9 | Rodrigues et al., 2018 |
| Faustovirus | Faustovirus mariensis | Urban lake water | Pampulha Lagoon (2019) | 466,080 | 483 | 0 | 36 | Borges et al., 2019 |
| Yaravirus | Yaravirus brasiliensis | Muddy water | Pampulha Lagoon (2020) | 44,924 | 74 | 68 (91.9%) | 57.9 | Boratto et al., 2020 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Boratto, P.V.M.; Serafim, M.S.M.; Witt, A.S.A.; Crispim, A.P.C.; Azevedo, B.L.d.; Souza, G.A.P.d.; Aquino, I.L.M.d.; Machado, T.B.; Queiroz, V.F.; Rodrigues, R.A.L.; et al. A Brief History of Giant Viruses’ Studies in Brazilian Biomes. Viruses 2022, 14, 191. https://0-doi-org.brum.beds.ac.uk/10.3390/v14020191
Boratto PVM, Serafim MSM, Witt ASA, Crispim APC, Azevedo BLd, Souza GAPd, Aquino ILMd, Machado TB, Queiroz VF, Rodrigues RAL, et al. A Brief History of Giant Viruses’ Studies in Brazilian Biomes. Viruses. 2022; 14(2):191. https://0-doi-org.brum.beds.ac.uk/10.3390/v14020191
Chicago/Turabian StyleBoratto, Paulo Victor M., Mateus Sá M. Serafim, Amanda Stéphanie A. Witt, Ana Paula C. Crispim, Bruna Luiza de Azevedo, Gabriel Augusto P. de Souza, Isabella Luiza M. de Aquino, Talita B. Machado, Victória F. Queiroz, Rodrigo A. L. Rodrigues, and et al. 2022. "A Brief History of Giant Viruses’ Studies in Brazilian Biomes" Viruses 14, no. 2: 191. https://0-doi-org.brum.beds.ac.uk/10.3390/v14020191