Remote Actuation of Apoptosis in Liver Cancer Cells via Magneto-Mechanical Modulation of Iron Oxide Nanoparticles
Abstract
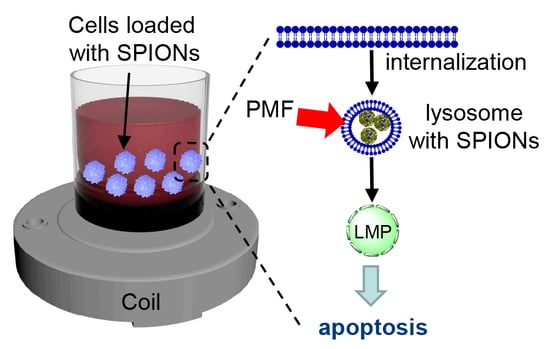
:1. Introduction
2. Results
2.1. Characterization of the Pulsed Magnetic Field (PMF) System and Internalization of SPIONs
2.2. PMF Stimulation of SPION-Loaded Cells Results in Cell Death
2.3. Triggering Apoptosis by PMF Stimulation via Disruption of Lysosomes
2.4. PMF Induces Apoptosis in 3D Multicellular Aggregates
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Cell Culture
4.3. Pulsed Magnetic Field (PMF) System Description and PMF Treatment
4.4. Cell Viability Assay
4.5. Detection of Intracellular Reactive Oxygen Species (ROS)
4.6. Caspase-3 Activity Assay
4.7. Detection of Apoptosis
4.8. Quantification of Mitochondrial Membrane Potential
4.9. Assessment of Lysosomal Integrity by Acridine Orange (AO)
4.10. Immunofluorescence Staining
4.11. Generation of 3D Multicellular Aggregates
4.12. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Pankhurst, Q.; Jones, S.; Dobson, J. Applications of magnetic nanoparticles in biomedicine: The story so far. J. Phys. D Appl. Phys. 2016, 49, 501002. [Google Scholar] [CrossRef]
- Dobson, J. Remote control of cellular behaviour with magnetic nanoparticles. Nat. Nanotechnol. 2008, 3, 139–143. [Google Scholar] [CrossRef] [PubMed]
- Gaster, R.S.; Hall, D.A.; Nielsen, C.H.; Osterfeld, S.J.; Yu, H.; Mach, K.E.; Wilson, R.J.; Murmann, B.; Liao, J.C.; Gambhir, S.S.; et al. Matrix-insensitive protein assays push the limits of biosensors in medicine. Nat. Med. 2009, 15, 1327–1332. [Google Scholar] [CrossRef] [PubMed]
- Haun, J.B.; Castro, C.M.; Wang, R.; Peterson, V.M.; Marinelli, B.S.; Lee, H.; Weissleder, R. Micro-NMR for rapid molecular analysis of human tumor samples. Sci. Transl. Med. 2011, 3, 71ra16. [Google Scholar] [CrossRef] [PubMed]
- Kircher, M.F.; Gambhir, S.S.; Grimm, J. Noninvasive cell-tracking methods. Nat. Rev. Clin. Oncol. 2011, 8, 677–688. [Google Scholar] [CrossRef]
- Kircher, M.F.; Mahmood, U.; King, R.S.; Weissleder, R.; Josephson, L. A multimodal nanoparticle for preoperative magnetic resonance imaging and intraoperative optical brain tumor delineation. Cancer Res. 2003, 63, 8122–8125. [Google Scholar]
- Kozissnik, B.; Bohorquez, A.C.; Dobson, J.; Rinaldi, C. Magnetic fluid hyperthermia: Advances, challenges, and opportunity. Int. J. Hyperth. 2013, 29, 706–714. [Google Scholar] [CrossRef]
- Guo, M.; Que, C.; Wang, C.; Liu, X.; Yan, H.; Liu, K. Multifunctional superparamagnetic nanocarriers with folate-mediated and pH-responsive targeting properties for anticancer drug delivery. Biomaterials 2011, 32, 185–194. [Google Scholar] [CrossRef]
- Golovin, Y.I.; Gribanovsky, S.L.; Golovin, D.Y.; Klyachko, N.L.; Majouga, A.G.; Master, A.M.; Sokolsky, M.; Kabanov, A.V. Towards nanomedicines of the future: Remote magneto-mechanical actuation of nanomedicines by alternating magnetic fields. J. Control. Release 2015, 219, 43–60. [Google Scholar] [CrossRef]
- Tseng, P.; Judy, J.W.; Di Carlo, D. Magnetic nanoparticle-mediated massively parallel mechanical modulation of single-cell behavior. Nat. Methods 2012, 9, 1113–1119. [Google Scholar] [CrossRef]
- Colombo, M.; Carregal-Romero, S.; Casula, M.F.; Gutierrez, L.; Morales, M.P.; Bohm, I.B.; Heverhagen, J.T.; Prosperi, D.; Parak, W.J. Biological applications of magnetic nanoparticles. Chem. Soc. Rev. 2012, 41, 4306–4334. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Rozhkova, E.A.; Ulasov, I.V.; Bader, S.D.; Rajh, T.; Lesniak, M.S.; Novosad, V. Biofunctionalized magnetic-vortex microdiscs for targeted cancer-cell destruction. Nat. Mater. 2010, 9, 165–171. [Google Scholar] [CrossRef] [PubMed]
- Master, A.M.; Williams, P.N.; Pothayee, N.; Pothayee, N.; Zhang, R.; Vishwasrao, H.M.; Golovin, Y.I.; Riffle, J.S.; Sokolsky, M.; Kabanov, A.V. Remote actuation of magnetic nanoparticles for cancer cell selective treatment through cytoskeletal disruption. Sci. Rep. 2016, 6, 33560. [Google Scholar] [CrossRef] [PubMed]
- Leulmi, S.; Chauchet, X.; Morcrette, M.; Ortiz, G.; Joisten, H.; Sabon, P.; Livache, T.; Hou, Y.X.; Carriere, M.; Lequien, S.; et al. Triggering the apoptosis of targeted human renal cancer cells by the vibration of anisotropic magnetic particles attached to the cell membrane. Nanoscale 2015, 7, 15904–15914. [Google Scholar] [CrossRef] [PubMed]
- Domenech, M.; Marrero-Berrios, I.; Torres-Lugo, M.; Rinaldi, C. Lysosomal membrane permeabilization by targeted magnetic nanoparticles in alternating magnetic fields. ACS Nano 2013, 7, 5091–5101. [Google Scholar] [CrossRef]
- Zhang, E.; Kircher, M.F.; Koch, M.; Eliasson, L.; Goldberg, S.N.; Renstrom, E. Dynamic magnetic fields remote-control apoptosis via nanoparticle rotation. ACS Nano 2014, 8, 3192–3201. [Google Scholar] [CrossRef]
- Wong, W.; Gan, W.L.; Liu, N.; Lew, W.S. Magneto-actuated cell apoptosis by biaxial pulsed magnetic field. Sci. Rep. 2017, 7, 10919. [Google Scholar] [CrossRef]
- Wong, W.; Gan, W.L.; Teo, Y.K.; Lew, W.S. Interplay of cell death signaling pathways mediated by alternating magnetic field gradient. Cell Death Discov. 2018, 4, 49. [Google Scholar] [CrossRef]
- Shin, Y.C.; Song, S.J.; Hong, S.W.; Jeong, S.J.; Chrzanowski, W.; Lee, J.C.; Han, D.W. Multifaceted biomedical applications of functional graphene nanomaterials to coated substrates, patterned arrays and hybrid scaffolds. Nanomaterials 2017, 7, 369. [Google Scholar] [CrossRef]
- Elbez, R.; McNaughton, B.H.; Patel, L.; Pienta, K.J.; Kopelman, R. Nanoparticle induced cell magneto-rotation: Monitoring morphology, stress and drug sensitivity of a suspended single cancer cell. PLoS ONE 2011, 6, e28475. [Google Scholar] [CrossRef]
- Hapuarachchige, S.; Kato, Y.; Ngen, E.J.; Smith, B.; Delannoy, M.; Artemov, D. Non-temperature induced effects of magnetized iron oxide nanoparticles in alternating magnetic field in cancer cells. PLoS ONE 2016, 11, e0156294. [Google Scholar] [CrossRef]
- Uzhytchak, M.; Lynnyk, A.; Zablotskii, V.; Dempsey, N.M.; Dias, A.L.; Bonfim, M.; Lunova, M.; Jirsa, M.; Kubinova, S.; Lunov, O.; et al. The use of pulsed magnetic fields to increase the uptake of iron oxide nanoparticles by living cells. Appl. Phys. Lett. 2017, 111, 243703. [Google Scholar] [CrossRef]
- Lunov, O.; Syrovets, T.; Rocker, C.; Tron, K.; Nienhaus, G.U.; Rasche, V.; Mailander, V.; Landfester, K.; Simmet, T. Lysosomal degradation of the carboxydextran shell of coated superparamagnetic iron oxide nanoparticles and the fate of professional phagocytes. Biomaterials 2010, 31, 9015–9022. [Google Scholar] [CrossRef]
- Lunov, O.; Syrovets, T.; Buchele, B.; Jiang, X.; Rocker, C.; Tron, K.; Nienhaus, G.U.; Walther, P.; Mailander, V.; Landfester, K.; et al. The effect of carboxydextran-coated superparamagnetic iron oxide nanoparticles on c-Jun N-terminal kinase-mediated apoptosis in human macrophages. Biomaterials 2010, 31, 5063–5071. [Google Scholar] [CrossRef]
- Tukmachev, D.; Lunov, O.; Zablotskii, V.; Dejneka, A.; Babic, M.; Sykova, E.; Kubinova, S. An effective strategy of magnetic stem cell delivery for spinal cord injury therapy. Nanoscale 2015, 7, 3954–3958. [Google Scholar] [CrossRef]
- Lunov, O.; Zablotskii, V.; Syrovets, T.; Rocker, C.; Tron, K.; Nienhaus, G.U.; Simmet, T. Modeling receptor-mediated endocytosis of polymer-functionalized iron oxide nanoparticles by human macrophages. Biomaterials 2011, 32, 547–555. [Google Scholar] [CrossRef]
- Zablotskii, V.; Lunov, O.; Dejneka, A.; Jastrabik, L.; Polyakova, T.; Syrovets, T.; Simmet, T. Nanomechanics of magnetically driven cellular endocytosis. Appl. Phys. Lett. 2011, 99, 183701. [Google Scholar] [CrossRef]
- Qin, J.; Laurent, S.; Jo, Y.S.; Roch, A.; Mikhaylova, M.; Bhujwalla, Z.M.; Muller, R.N.; Muhammed, M. A high-performance magnetic resonance Imaging T-2 contrast agent. Adv. Mater. 2007, 19, 2411. [Google Scholar] [CrossRef]
- Wang, Y.X.J.; Hussain, S.M.; Krestin, G.P. Superparamagnetic iron oxide contrast agents: Physicochemical characteristics and applications in MR imaging. Eur. Radiol. 2001, 11, 2319–2331. [Google Scholar] [CrossRef]
- Allkemper, T.; Bremer, C.; Matuszewski, L.; Ebert, W.; Reimer, P. Contrast-enhanced blood-pool MR angiography with optimized iron oxides: Effect of size and dose on vascular contrast enhancement in rabbits. Radiology 2002, 223, 432–438. [Google Scholar] [CrossRef]
- Wilhelm, S.; Tavares, A.J.; Dai, Q.; Ohta, S.; Audet, J.; Dvorak, H.F.; Chan, W.C.W. Analysis of nanoparticle delivery to tumours. Nat. Rev. Mater. 2016, 1, 16014. [Google Scholar] [CrossRef]
- Yu, M.; Zheng, J. Clearance pathways and tumor targeting of imaging nanoparticles. ACS Nano 2015, 9, 6655–6674. [Google Scholar] [CrossRef] [Green Version]
- Letters, A.P. The dose makes the poison. Nat. Nanotechnol. 2011, 6, 329. [Google Scholar] [CrossRef]
- Oberdorster, G. Safety assessment for nanotechnology and nanomedicine: Concepts of nanotoxicology. J. Intern. Med. 2010, 267, 89–105. [Google Scholar] [CrossRef]
- Feng, Q.; Liu, Y.; Huang, J.; Chen, K.; Huang, J.; Xiao, K. Uptake, distribution, clearance, and toxicity of iron oxide nanoparticles with different sizes and coatings. Sci. Rep. 2018, 8, 2082. [Google Scholar] [CrossRef]
- Hamm, B.; Staks, T.; Taupitz, M.; Maibauer, R.; Speidel, A.; Huppertz, A.; Frenzel, T.; Lawaczeck, R.; Wolf, K.J.; Lange, L. Contrast-enhanced MR imaging of liver and spleen: First experience in humans with a new superparamagnetic iron oxide. J. Magn. Reson. Imaging 1994, 4, 659–668. [Google Scholar] [CrossRef]
- Kemp, S.J.; Ferguson, R.M.; Khandhar, A.P.; Krishnan, K.M. Monodisperse magnetite nanoparticles with nearly ideal saturation magnetization. RSC Adv. 2016, 6, 77452–77464. [Google Scholar] [CrossRef]
- Zablotskii, V.; Polyakova, T.; Lunov, O.; Dejneka, A. How a high-gradient magnetic field could affect cell life. Sci. Rep. 2016, 6, 37407. [Google Scholar] [CrossRef]
- Zhu, W.; von dem Bussche, A.; Yi, X.; Qiu, Y.; Wang, Z.; Weston, P.; Hurt, R.H.; Kane, A.B.; Gao, H. Nanomechanical mechanism for lipid bilayer damage induced by carbon nanotubes confined in intracellular vesicles. Proc. Natl. Acad. Sci. USA 2016, 113, 12374–12379. [Google Scholar] [CrossRef] [Green Version]
- Kalwarczyk, T.; Ziebacz, N.; Bielejewska, A.; Zaboklicka, E.; Koynov, K.; Szymanski, J.; Wilk, A.; Patkowski, A.; Gapinski, J.; Butt, H.J.; et al. Comparative analysis of viscosity of complex liquids and cytoplasm of mammalian cells at the nanoscale. Nano Lett. 2011, 11, 2157–2163. [Google Scholar] [CrossRef]
- Kast, D.J.; Dominguez, R. The cytoskeleton-autophagy connection. Curr. Biol. 2017, 27, R318–R326. [Google Scholar] [CrossRef] [Green Version]
- Pu, J.; Guardia, C.M.; Keren-Kaplan, T.; Bonifacino, J.S. Mechanisms and functions of lysosome positioning. J. Cell Sci. 2016, 129, 4329–4339. [Google Scholar] [CrossRef] [Green Version]
- Schwake, M.; Schroder, B.; Saftig, P. Lysosomal membrane proteins and their central role in physiology. Traffic 2013, 14, 739–748. [Google Scholar] [CrossRef]
- Gonzalez-Rodriguez, D.; Guillou, L.; Cornat, F.; Lafaurie-Janvore, J.; Babataheri, A.; de Langre, E.; Barakat, A.I.; Husson, J. Mechanical criterion for the rupture of a cell membrane under compression. Biophys. J. 2016, 111, 2711–2721. [Google Scholar] [CrossRef] [Green Version]
- Pierzynska-Mach, A.; Janowski, P.A.; Dobrucki, J.W. Evaluation of acridine orange, lysotracker red, and quinacrine as fluorescent probes for long-term tracking of acidic vesicles. Cytom. Part A 2014, 85, 729–737. [Google Scholar] [CrossRef]
- Kirkegaard, T.; Roth, A.G.; Petersen, N.H.T.; Mahalka, A.K.; Olsen, O.D.; Moilanen, I.; Zylicz, A.; Knudsen, J.; Sandhoff, K.; Arenz, C.; et al. Hsp70 stabilizes lysosomes and reverts Niemann-Pick disease-associated lysosomal pathology. Nature 2010, 463, 549–553. [Google Scholar] [CrossRef]
- Petersen, N.H.T.; Olsen, O.D.; Groth-Pedersen, L.; Ellegaard, A.M.; Bilgin, M.; Redmer, S.; Ostenfeld, M.S.; Ulanet, D.; Dovmark, T.H.; Lonborg, A.; et al. Transformation-associated changes in sphingolipid metabolism sensitize cells to lysosomal cell death induced by inhibitors of acid sphingomyelinase. Cancer Cell 2013, 24, 379–393. [Google Scholar] [CrossRef] [Green Version]
- Boya, P.; Kroemer, G. Lysosomal membrane permeabilization in cell death. Oncogene 2008, 27, 6434–6451. [Google Scholar] [CrossRef] [Green Version]
- Settembre, C.; Fraldi, A.; Medina, D.L.; Ballabio, A. Signals from the lysosome: A control centre for cellular clearance and energy metabolism. Nat. Rev. Mol. Cell Biol. 2013, 14, 283–296. [Google Scholar] [CrossRef] [Green Version]
- Boya, P.; Andreau, K.; Poncet, D.; Zamzami, N.; Perfettini, J.L.; Metivier, D.; Ojcius, D.M.; Jaattela, M.; Kroemer, G. Lysosomal membrane permeabilization induces cell death in a mitochondrion-dependent fashion. J. Exp. Med. 2003, 197, 1323–1334. [Google Scholar] [CrossRef]
- Erdal, H.; Berndtsson, M.; Castro, J.; Brunk, U.; Shoshan, M.C.; Linder, S. Induction of lysosomal membrane permeabilization by compounds that activate p53-independent apoptosis. Proc. Natl. Acad. Sci. USA 2005, 102, 192–197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lynnyk, A.; Lunova, M.; Jirsa, M.; Egorova, D.; Kulikov, A.; Kubinova, S.; Lunov, O.; Dejneka, A. Manipulating the mitochondria activity in human hepatic cell line Huh7 by low-power laser irradiation. Biomed. Opt. Express 2018, 9, 1283–1300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smolkova, B.; Lunova, M.; Lynnyk, A.; Uzhytchak, M.; Churpita, O.; Jirsa, M.; Kubinova, S.; Lunov, O.; Dejneka, A. Non-thermal plasma, as a new physicochemical source, to induce redox imbalance and subsequent cell death in liver cancer cell lines. Cell. Physiol. Biochem. 2019, 52, 119–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lunova, M.; Prokhorov, A.; Jirsa, M.; Hof, M.; Olzynska, A.; Jurkiewicz, P.; Kubinova, S.; Lunov, O.; Dejneka, A. Nanoparticle core stability and surface functionalization drive the mTOR signaling pathway in hepatocellular cell lines. Sci. Rep. 2017, 7, 16049. [Google Scholar] [CrossRef] [Green Version]
- Krysko, O.; de Ridder, L.; Cornelissen, M. Phosphatidylserine exposure during early primary necrosis (oncosis) in JB6 cells as evidenced by immunogold labeling technique. Apoptosis 2004, 9, 495–500. [Google Scholar] [CrossRef]
- Galluzzi, L.; Vitale, I.; Aaronson, S.A.; Abrams, J.M.; Adam, D.; Agostinis, P.; Alnemri, E.S.; Altucci, L.; Amelio, I.; Andrews, D.W.; et al. Molecular mechanisms of cell death: Recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ. 2018, 25, 486–541. [Google Scholar] [CrossRef]
- Galluzzi, L.; Vitale, I.; Abrams, J.M.; Alnemri, E.S.; Baehrecke, E.H.; Blagosklonny, M.V.; Dawson, T.M.; Dawson, V.L.; El-Deiry, W.S.; Fulda, S.; et al. Molecular definitions of cell death subroutines: Recommendations of the Nomenclature Committee on Cell Death 2012. Cell Death Differ. 2012, 19, 107–120. [Google Scholar] [CrossRef]
- Kroemer, G.; Galluzzi, L.; Vandenabeele, P.; Abrams, J.; Alnemri, E.S.; Baehrecke, E.H.; Blagosklonny, M.V.; El-Deiry, W.S.; Golstein, P.; Green, D.R.; et al. Classification of cell death: Recommendations of the Nomenclature Committee on Cell Death 2009. Cell Death Differ. 2009, 16, 3–11. [Google Scholar] [CrossRef]
- Lunov, O.; Zablotskii, V.; Churpita, O.; Lunova, M.; Jirsa, M.; Dejneka, A.; Kubinova, S. Chemically different non-thermal plasmas target distinct cell death pathways. Sci. Rep. 2017, 7, 600. [Google Scholar] [CrossRef] [Green Version]
- Kang, M.A.; So, E.Y.; Simons, A.L.; Spitz, D.R.; Ouchi, T. DNA damage induces reactive oxygen species generation through the H2AX-Nox1/Rac1 pathway. Cell Death Dis. 2012, 3, e249. [Google Scholar] [CrossRef]
- Lunov, O.; Zablotskii, V.; Churpita, O.; Jager, A.; Polivka, L.; Sykova, E.; Dejneka, A.; Kubinova, S. The interplay between biological and physical scenarios of bacterial death induced by non-thermal plasma. Biomaterials 2016, 82, 71–83. [Google Scholar] [CrossRef] [PubMed]
- Lunova, M.; Smolkova, B.; Uzhytchak, M.; Janouskova, K.Z.; Jirsa, M.; Egorova, D.; Kulikov, A.; Kubinova, S.; Dejneka, A.; Lunov, O. Light-induced modulation of the mitochondrial respiratory chain activity: Possibilities and limitations. Cell Mol. Life Sci. 2019. [Google Scholar] [CrossRef] [PubMed]
- Langhans, S.A. Three-dimensional in vitro cell culture models in drug discovery and drug repositioning. Front. Pharmacol. 2018, 9, 6. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Farach-Carson, M.C.; Jia, X. Three-dimensional in vitro tumor models for cancer research and drug evaluation. Biotechnol. Adv. 2014, 32, 1256–1268. [Google Scholar] [CrossRef] [Green Version]
- Tomasini, M.D.; Rinaldi, C.; Tomassone, M.S. Molecular dynamics simulations of rupture in lipid bilayers. Exp. Biol. Med. 2010, 235, 181–188. [Google Scholar] [CrossRef]
- Hare, J.I.; Lammers, T.; Ashford, M.B.; Puri, S.; Storm, G.; Barry, S.T. Challenges and strategies in anti-cancer nanomedicine development: An industry perspective. Adv. Drug Deliv. Rev. 2017, 108, 25–38. [Google Scholar] [CrossRef] [Green Version]
- Hua, S.; de Matos, M.B.C.; Metselaar, J.M.; Storm, G. Current trends and challenges in the clinical translation of nanoparticulate nanomedicines: Pathways for translational development and commercialization. Front. Pharmacol. 2018, 9, 790. [Google Scholar] [CrossRef]
- Rosenblum, D.; Joshi, N.; Tao, W.; Karp, J.M.; Peer, D. Progress and challenges towards targeted delivery of cancer therapeutics. Nat. Commun. 2018, 9, 1410. [Google Scholar] [CrossRef] [Green Version]
- Barenholz, Y. Doxil(R)--the first FDA-approved nano-drug: Lessons learned. J. Control. Release 2012, 160, 117–134. [Google Scholar] [CrossRef]
- Zhang, Y.N.; Poon, W.; Tavares, A.J.; McGilvray, I.D.; Chan, W.C.W. Nanoparticle-liver interactions: Cellular uptake and hepatobiliary elimination. J. Control. Release 2016, 240, 332–348. [Google Scholar] [CrossRef]
- Piao, S.; Amaravadi, R.K. Targeting the lysosome in cancer. Ann. N. Y. Acad. Sci. 2016, 1371, 45–54. [Google Scholar] [CrossRef] [Green Version]
- Serrano-Puebla, A.; Boya, P. Lysosomal membrane permeabilization as a cell death mechanism in cancer cells. Biochem. Soc. Trans. 2018, 46, 207–215. [Google Scholar] [CrossRef]
- Joris, F.; De Backer, L.; Van de Vyver, T.; Bastiancich, C.; De Smedt, S.C.; Raemdonck, K. Repurposing cationic amphiphilic drugs as adjuvants to induce lysosomal siRNA escape in nanogel transfected cells. J. Control. Release 2018, 269, 266–276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lunova, M.; Smolkova, B.; Lynnyk, A.; Uzhytchak, M.; Jirsa, M.; Kubinova, S.; Dejneka, A.; Lunov, O. Targeting the mTOR signaling pathway utilizing nanoparticles: A critical overview. Cancers 2019, 11, 82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fehrenbacher, N.; Jäättelä, M. Lysosomes as targets for cancer therapy. Cancer Res. 2005, 65, 2993–2995. [Google Scholar] [CrossRef]
- Wang, F.; Gomez-Sintes, R.; Boya, P. Lysosomal membrane permeabilization and cell death. Traffic 2018, 19, 918–931. [Google Scholar] [CrossRef]
- Ni, Z.H.; Wang, B.; Dai, X.F.; Ding, W.; Yang, T.; Li, X.Z.; Lewin, S.; Xu, L.; Lian, J.Q.; He, F.T. HCC cells with high levels of Bcl-2 are resistant to ABT-737 via activation of the ROS-JNK-autophagy pathway. Free Radic. Biol. Med. 2014, 70, 194–203. [Google Scholar] [CrossRef]
- Guo, L.; Dial, S.; Shi, L.M.; Branham, W.; Liu, J.; Fang, J.L.; Green, B.; Deng, H.; Kaput, J.; Ning, B.T. Similarities and differences in the expression of drug-metabolizing enzymes between human hepatic cell lines and primary human hepatocytes. Drug Metab. Dispos. 2011, 39, 528–538. [Google Scholar] [CrossRef] [Green Version]
- Pattingre, S.; Levine, B. Bcl-2 inhibition of autophagy: A new route to cancer? Cancer Res. 2006, 66, 2885–2888. [Google Scholar] [CrossRef]
- Lindqvist, L.M.; Heinlein, M.; Huang, D.C.S.; Vaux, D.L. Prosurvival Bcl-2 family members affect autophagy only indirectly, by inhibiting Bax and Bak. Proc. Natl. Acad. Sci. USA 2014, 111, 8512–8517. [Google Scholar] [CrossRef] [Green Version]
- Ashkenazi, A.; Fairbrother, W.J.; Leverson, J.D.; Souers, A.J. From basic apoptosis discoveries to advanced selective BCL-2 family inhibitors. Nat. Rev. Drug Discov. 2017, 16, 273–284. [Google Scholar] [CrossRef] [PubMed]
- Lessene, G.; Czabotar, P.E.; Colman, P.M. BCL-2 family antagonists for cancer therapy. Nat. Rev. Drug Discov. 2008, 7, 989–1000. [Google Scholar] [CrossRef] [PubMed]
- Johansson, A.C.; Appelqvist, H.; Nilsson, C.; Kagedal, K.; Roberg, K.; Ollinger, K. Regulation of apoptosis-associated lysosomal membrane permeabilization. Apoptosis 2010, 15, 527–540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mrschtik, M.; Ryan, K.M. Lysosomal proteins in cell death and autophagy. FEBS J. 2015, 282, 1858–1870. [Google Scholar] [CrossRef] [PubMed]
- Lunov, O.; Zablotskii, V.; Churpita, O.; Chanova, E.; Sykova, E.; Dejneka, A.; Kubinova, S. Cell death induced by ozone and various non-thermal plasmas: Therapeutic perspectives and limitations. Sci. Rep. 2014, 4, 7129. [Google Scholar] [CrossRef] [Green Version]
- Smiley, S.T.; Reers, M.; Mottola-Hartshorn, C.; Lin, M.; Chen, A.; Smith, T.W.; Steele, G.D., Jr.; Chen, L.B. Intracellular heterogeneity in mitochondrial membrane potentials revealed by a J-aggregate-forming lipophilic cation JC-1. Proc. Natl. Acad. Sci. USA 1991, 88, 3671–3675. [Google Scholar] [CrossRef] [Green Version]
- Zuliani, T.; Duval, R.; Jayat, C.; Schnebert, S.; Andre, P.; Dumas, M.; Ratinaud, M.H. Sensitive and reliable JC-1 and TOTO-3 double staining to assess mitochondrial transmembrane potential and plasma membrane integrity: Interest for cell death investigations. Cytom. A 2003, 54, 100–108. [Google Scholar] [CrossRef]
- Foty, R. A simple hanging drop cell culture protocol for generation of 3D spheroids. J. Vis. Exp. 2011. [Google Scholar] [CrossRef]
- Petrenko, Y.; Sykova, E.; Kubinova, S. The therapeutic potential of three-dimensional multipotent mesenchymal stromal cell spheroids. Stem Cell Res. Ther. 2017, 8, 94. [Google Scholar] [CrossRef] [Green Version]
- Hamilton, N. Quantification and its applications in fluorescent microscopy imaging. Traffic 2009, 10, 951–961. [Google Scholar] [CrossRef]
- Dell, R.B.; Holleran, S.; Ramakrishnan, R. Sample size determination. ILAR J. 2002, 43, 207–213. [Google Scholar] [CrossRef] [PubMed]





© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lunov, O.; Uzhytchak, M.; Smolková, B.; Lunova, M.; Jirsa, M.; Dempsey, N.M.; Dias, A.L.; Bonfim, M.; Hof, M.; Jurkiewicz, P.; et al. Remote Actuation of Apoptosis in Liver Cancer Cells via Magneto-Mechanical Modulation of Iron Oxide Nanoparticles. Cancers 2019, 11, 1873. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers11121873
Lunov O, Uzhytchak M, Smolková B, Lunova M, Jirsa M, Dempsey NM, Dias AL, Bonfim M, Hof M, Jurkiewicz P, et al. Remote Actuation of Apoptosis in Liver Cancer Cells via Magneto-Mechanical Modulation of Iron Oxide Nanoparticles. Cancers. 2019; 11(12):1873. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers11121873
Chicago/Turabian StyleLunov, Oleg, Mariia Uzhytchak, Barbora Smolková, Mariia Lunova, Milan Jirsa, Nora M. Dempsey, André L. Dias, Marlio Bonfim, Martin Hof, Piotr Jurkiewicz, and et al. 2019. "Remote Actuation of Apoptosis in Liver Cancer Cells via Magneto-Mechanical Modulation of Iron Oxide Nanoparticles" Cancers 11, no. 12: 1873. https://0-doi-org.brum.beds.ac.uk/10.3390/cancers11121873