One Pot Use of Combilipases for Full Modification of Oils and Fats: Multifunctional and Heterogeneous Substrates
Abstract
:1. Introduction
1.1. Enzymatic Biocatalysis
1.2. Enzymatic Full Modification of the Substrate
1.2.1. Modification of Monofunctional Substrates
1.2.2. Modification of Multifunctional Substrates
1.3. Lipases as Industrial Biocatalysts
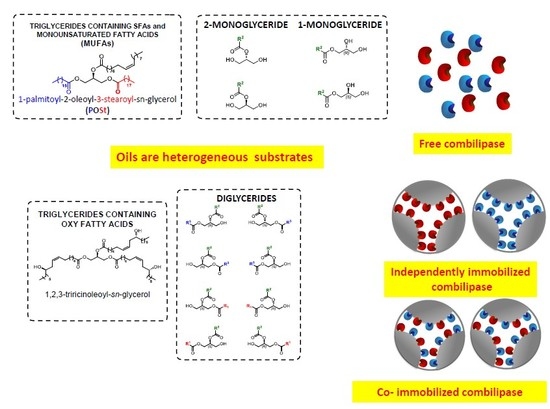
2. Oils and Fats as Heterogeneous Substrates
2.1. Lipase Production of Free Fatty Acids via Hydrolysis of Oils and Fats
2.2. Biodiesel Production Using Lipases as Biocatalysts
2.2.1. Transesterification
2.2.2. Hydroesterification
3. Advantages of the Simultaneous Use of Several Lipases to Fully Modify Multifunctional Substrates: The Concept of Combilipases
4. Use of Lipases from Microorganisms that Produce Several Lipases
5. Use of Lipases Mixtures in Liquid Formulations
5.1. Hydrolysis of Oils and Fats Using Combilipases in Liquid Form
5.2. Use of Combilipases in Liquid Form in Biodiesel Production
5.3. Other Uses of Combilipases in Liquid Formulations
6. Use of Individually Immobilized Lipases
6.1. Use of Mixtures of Immobilized Lipases
6.1.1. Hydrolysis of Oils and Fats Using Individually Immobilized Combilipases
6.1.2. Use of Individually Immobilized Combilipases in Biodiesel Production
6.1.3. Other Uses of Immobilized Combilipases
6.2. Use of Mixtures of the Same Lipase Immobilized Following Different Protocols: A Special Combilipase
7. Use of Coimmobilized Lipases
7.1. Coimmobilization of Lipases: Advantages, Problems and Proposed Solutions
7.2. Use of Coimmobilized Combilipases in Biodiesel Production
7.3. Preparation of Coimmobilized Combilipases to Reuse the Most Stable Enzymes
8. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Chapman, J.; Ismail, A.; Dinu, C. Industrial applications of enzymes: Recent advances, techniques, and outlooks. Catalysts 2018, 8, 238. [Google Scholar] [CrossRef] [Green Version]
- De Gonzalo, G.; Domínguez de María, P. Biocatalysis: An industrial perspective. In Catalysis Series; de Gonzalo, G., Domínguez de María, P., Eds.; Royal Society of Chemistry: Cambridge, UK, 2017; pp. 1–511. ISBN 978-1-78262-619-0. [Google Scholar]
- Hughes, D.L. Biocatalysis in drug development-highlights of the recent patent literature. Org. Process Res. Dev. 2018, 22, 1063–1080. [Google Scholar] [CrossRef]
- Li, G.; Wang, J.; Reetz, M.T. Biocatalysts for the pharmaceutical industry created by structure-guided directed evolution of stereoselective enzymes. Bioorg. Med. Chem. 2018, 26, 1241–1251. [Google Scholar] [CrossRef] [PubMed]
- Raveendran, S.; Parameswaran, B.; Ummalyma, S.B.; Abraham, A.; Mathew, A.K.; Madhavan, A.; Rebello, S.; Pandey, A. Applications of microbial enzymes in food industry. Food Technol. Biotechnol. 2018, 56, 16–30. [Google Scholar] [CrossRef] [PubMed]
- Rosenthal, K.; Lütz, S. Recent developments and challenges of biocatalytic processes in the pharmaceutical industry. Curr. Opin. Green Sustain. Chem. 2018, 11, 58–64. [Google Scholar] [CrossRef]
- Abdelraheem, E.M.M.; Busch, H.; Hanefeld, U.; Tonin, F. Biocatalysis explained: From pharmaceutical to bulk chemical production. React. Chem. Eng. 2019, 4, 1878–1894. [Google Scholar] [CrossRef] [Green Version]
- Adams, J.P.; Brown, M.J.B.; Diaz-Rodriguez, A.; Lloyd, R.C.; Roiban, G.-D. Biocatalysis: A pharma perspective. Adv. Synth. Catal. 2019, 361, 2421–2432. [Google Scholar] [CrossRef] [Green Version]
- Basso, A.; Serban, S. Industrial applications of immobilized enzymes—A review. Mol. Catal. 2019, 479, 110607. [Google Scholar] [CrossRef]
- Bilal, M.; Iqbal, H.M.N. Sustainable bioconversion of food waste into high-value products by immobilized enzymes to meet bio-economy challenges and opportunities—A review. Food Res. Int. 2019, 123, 226–240. [Google Scholar] [CrossRef]
- Bilal, M.; Iqbal, H.M.N. State-of-the-art strategies and applied perspectives of enzyme biocatalysis in food sector—Current status and future trends. Crit. Rev. Food Sci. Nutr. 2019, 1–15. [Google Scholar] [CrossRef]
- Faber, K.; Fessner, W.-D.; Turner, N.J. Biocatalysis: Ready to master increasing complexity. Adv. Synth. Catal. 2019, 361, 2373–2376. [Google Scholar] [CrossRef] [Green Version]
- Domínguez de María, P.; de Gonzalo, G.; Alcántara, A.R. Biocatalysis as useful tool in asymmetric synthesis: An assessment of recently granted patents (2014–2019). Catalysts 2019, 9, 802. [Google Scholar] [CrossRef] [Green Version]
- Sheldon, R.A.; Brady, D.; Bode, M.L. The Hitchhiker’s guide to biocatalysis: Recent advances in the use of enzymes in organic synthesis. Chem. Sci. 2020, 11, 2587–2605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Foley, A.M.; Maguire, A.R. The impact of recent developments in technologies which enable the increased use of biocatalysts. Eur. J. Org. Chem. 2019, 2019, 3713–3734. [Google Scholar] [CrossRef]
- Guajardo, N.; Domínguez de María, P. Continuous biocatalysis in environmentally-friendly media: A triple synergy for future sustainable processes. ChemCatChem 2019, 11, 3128–3137. [Google Scholar] [CrossRef]
- Prier, C.K.; Kosjek, B. Recent preparative applications of redox enzymes. Curr. Opin. Chem. Biol. 2019, 49, 105–112. [Google Scholar] [CrossRef]
- Sheldon, R.A.; Brady, D. Broadening the scope of biocatalysis in sustainable organic synthesis. ChemSusChem 2019, 12, 2859–2881. [Google Scholar] [CrossRef]
- Woodley, J.M. Accelerating the implementation of biocatalysis in industry. Appl. Microbiol. Biotechnol. 2019, 103, 4733–4739. [Google Scholar] [CrossRef]
- Hoyos, P.; Pace, V.; Hernaiz, M.; Alcántara, A.R. Biocatalysis in the pharmaceutical industry. A greener future. Curr. Green Chem. 2014, 1, 155–181. [Google Scholar] [CrossRef]
- Alcántara, A.R. Biotransformations in Drug synthesis: A Green and powerful tool for medicinal chemistry. J. Med. Chem. Drug Des. 2018, 1, 1–7. [Google Scholar]
- Alcántara, A.R. Biocatalysis and pharmaceuticals: A smart tool for sustainable development. Catalysts 2019, 9, 792. [Google Scholar] [CrossRef] [Green Version]
- Sheldon, R.A.; Woodley, J.M. Role of biocatalysis in sustainable chemistry. Chem. Rev. 2018, 118, 801–838. [Google Scholar] [CrossRef] [PubMed]
- Sheldon, R.A. Biocatalysis and green chemistry. In Green Biocatalysis; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2016; pp. 1–15. ISBN 9781118828083. [Google Scholar]
- Woodley, J.M. New frontiers in biocatalysis for sustainable synthesis. Curr. Opin. Green Sustain. Chem. 2020, 21, 22–26. [Google Scholar] [CrossRef]
- Schoemaker, H.E. Dispelling the myths - Biocatalysis in industrial synthesis. Science 2003, 299, 1694–1697. [Google Scholar] [CrossRef] [PubMed]
- Ferrer, M.; Beloqui, A.; Timmis, K.; Golyshin, P. Metagenomics for mining new genetic resources of microbial communities. J. Mol. Microbiol. Biotechnol. 2009, 16, 109–123. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Arrojo, L.; Guazzaroni, M.-E.; López-Cortés, N.; Beloqui, A.; Ferrer, M. Metagenomic era for biocatalyst identification. Curr. Opin. Biotechnol. 2010, 21, 725–733. [Google Scholar] [CrossRef]
- Ferrer, M.; Martínez-Martínez, M.; Bargiela, R.; Streit, W.R.; Golyshina, O.V.; Golyshin, P.N. Estimating the success of enzyme bioprospecting through metagenomics: Current status and future trends. Microb. Biotechnol. 2016, 9, 22–34. [Google Scholar] [CrossRef]
- Vieites, J.M.; Guazzaroni, M.-E.; Beloqui, A.; Golyshin, P.N.; Ferrer, M. Metagenomics approaches in systems microbiology. FEMS Microbiol. Rev. 2009, 33, 236–255. [Google Scholar] [CrossRef]
- Guazzaroni, M.E.; Beloqui, A.; Golyshin, P.N.; Ferrer, M. Metagenomics as a new technological tool to gain scientific knowledge. World J. Microbiol. Biotechnol. 2009, 25, 945–954. [Google Scholar] [CrossRef]
- Bilal, M.; Iqbal, H.M.N. Tailoring multipurpose biocatalysts via protein engineering approaches: A review. Catal. Letters 2019, 149, 2204–2217. [Google Scholar] [CrossRef]
- Steiner, K.; Schwab, H. Recent advances in rational approaches for enzyme engineering. Comput. Struct. Biotechnol. J. 2012, 2, e201209010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Porebski, B.T.; Buckle, A.M. Consensus protein design. Protein Eng. Des. Sel. 2016, 29, 245–251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Q.; Xiao, Y.; Zhang, W.; Mu, W. Current methods and applications in computational protein design for food industry. Crit. Rev. Food Sci. Nutr. 2019. In press. [Google Scholar] [CrossRef] [PubMed]
- Wilding, M.; Hong, N.; Spence, M.; Buckle, A.M.; Jackson, C.J. Protein engineering: The potential of remote mutations. Biochem. Soc. Trans. 2019, 47, 701–711. [Google Scholar] [CrossRef] [PubMed]
- Musil, M.; Konegger, H.; Hon, J.; Bednar, D.; Damborsky, J. Computational design of stable and soluble biocatalysts. ACS Catal. 2019, 9, 1033–1054. [Google Scholar] [CrossRef] [Green Version]
- Bornscheuer, U.T.; Hauer, B.; Jaeger, K.E.; Schwaneberg, U. Directed evolution empowered redesign of natural proteins for the sustainable production of chemicals and pharmaceuticals. Angew. Chemie Int. Ed. 2019, 58, 36–40. [Google Scholar] [CrossRef]
- Zeymer, C.; Hilvert, D. Directed evolution of protein catalysts. Annu. Rev. Biochem. 2018, 87, 131–157. [Google Scholar] [CrossRef]
- Arnold, F.H. Directed evolution: Bringing new chemistry to life. Angew. Chemie Int. Ed. 2018, 57, 4143–4148. [Google Scholar] [CrossRef] [Green Version]
- Reetz, M.T. Directed Evolution of Selective Enzymes: Catalysts for Organic Chemistry and Biotechnology; Wiley-VCH Verlag GmbH & Co. KGaA: Weinheim, Germany, 2016; ISBN 3527655484. [Google Scholar]
- Qu, G.; Li, A.; Acevedo-Rocha, C.G.; Sun, Z.; Reetz, M.T. The crucial role of methodology development in directed evolution of selective enzymes. Angew. Chemie Int. Ed. 2019. [Google Scholar] [CrossRef]
- Markel, U.; Essani, K.D.; Besirlioglu, V.; Schiffels, J.; Streit, W.R.; Schwaneberg, U. Advances in ultrahigh-throughput screening for directed enzyme evolution. Chem. Soc. Rev. 2020, 49, 233–262. [Google Scholar] [CrossRef]
- Sayous, V.; Lubrano, P.; Li, Y.; Acevedo-Rocha, C.G. Unbiased libraries in protein directed evolution. Biochim. Biophys. Acta Proteins Proteom. 2020, 1868, 140321. [Google Scholar] [CrossRef] [PubMed]
- Arnold, F.H. Innovation by evolution: Bringing new chemistry to life (Nobel lecture). Angew. Chemie Int. Ed. 2019, 58, 14420–14426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baker, D. What has de novo protein design taught us about protein folding and biophysics? Protein Sci. 2019, 28, 678–683. [Google Scholar] [CrossRef] [Green Version]
- Chowdhury, R.; Maranas, C.D. From directed evolution to computational enzyme engineering—A review. AIChE J. 2020, 66, e16847. [Google Scholar] [CrossRef]
- Huang, P.-S.; Boyken, S.E.; Baker, D. The coming of age of de novo protein design. Nature 2016, 537, 320–327. [Google Scholar] [CrossRef]
- Korendovych, I.V.; DeGrado, W.F. De novo protein design, a retrospective. Q. Rev. Biophys. 2020, 53, e3. [Google Scholar] [CrossRef]
- Mazurenko, S.; Prokop, Z.; Damborsky, J. Machine learning in enzyme engineering. ACS Catal. 2020, 10, 1210–1223. [Google Scholar] [CrossRef]
- Eraslan, G.; Avsec, Ž.; Gagneur, J.; Theis, F.J. Deep learning: New computational modelling techniques for genomics. Nat. Rev. Genet. 2019, 20, 389–403. [Google Scholar] [CrossRef]
- Han, X.; Wang, X.; Zhou, K. Develop machine learning-based regression predictive models for engineering protein solubility. Bioinformatics 2019, 35, 4640–4646. [Google Scholar] [CrossRef]
- Li, G.; Dong, Y.; Reetz, M.T. Can machine learning revolutionize directed evolution of selective enzymes? Adv. Synth. Catal. 2019, 361, 2377–2386. [Google Scholar] [CrossRef] [Green Version]
- DeSantis, G.; Jones, J.B. Chemical modification of enzymes for enhanced functionality. Curr. Opin. Biotechnol. 1999, 10, 324–330. [Google Scholar] [CrossRef]
- Cowan, D.A.; Fernandez-Lafuente, R. Enhancing the functional properties of thermophilic enzymes by chemical modification and immobilization. Enzyme Microb. Technol. 2011, 49, 326–346. [Google Scholar] [CrossRef] [PubMed]
- Davis, B.G. Chemical modification of biocatalysts. Curr. Opin. Biotechnol. 2003, 14, 379–386. [Google Scholar] [CrossRef]
- Díaz-Rodríguez, A.; Davis, B.G. Chemical modification in the creation of novel biocatalysts. Curr. Opin. Chem. Biol. 2011, 15, 211–219. [Google Scholar] [CrossRef]
- Thompson, M.P.; Derrington, S.R.; Heath, R.S.; Porter, J.L.; Mangas-Sanchez, J.; Devine, P.N.; Truppo, M.D.; Turner, N.J. A generic platform for the immobilisation of engineered biocatalysts. Tetrahedron 2019, 75, 327–334. [Google Scholar] [CrossRef]
- Ahmad, A.; Javed, M.R.; Ibrahim, M.; Sajid, A.; Hussain, K.; Kaleem, M.; Fatima, H.M.; Nadeem, H. Methods of enzyme immobilization on various supports. In Enzymatic Fuel Cells: Materials and Applications; Inamuddin, Ahmer, M.F., Ahamed, M.I., Asiri, A.M., Eds.; Materials Research Forum: Millersville, PA, USA, 2019; pp. 1–28. [Google Scholar]
- Garcia-Galan, C.; Berenguer-Murcia, Á.; Fernandez-Lafuente, R.; Rodrigues, R.C. Potential of different enzyme immobilization strategies to improve enzyme performance. Adv. Synth. Catal. 2011, 353, 2885–2904. [Google Scholar] [CrossRef]
- Rodrigues, R.C.; Ortiz, C.; Berenguer-Murcia, Á.; Torres, R.; Fernández-Lafuente, R. Modifying enzyme activity and selectivity by immobilization. Chem. Soc. Rev. 2013, 42, 6290–6307. [Google Scholar] [CrossRef]
- Mateo, C.; Palomo, J.M.; Fernandez-Lorente, G.; Guisan, J.M.; Fernandez-Lafuente, R. Improvement of enzyme activity, stability and selectivity via immobilization techniques. Enzyme Microb. Technol. 2007, 40, 1451–1463. [Google Scholar] [CrossRef]
- Sheldon, R.A.; van Pelt, S. Enzyme immobilization in biocatalysis: Why, what and how. Chem. Soc. Rev. 2013, 42, 6223–6235. [Google Scholar] [CrossRef] [Green Version]
- Fernandez-Lafuente, R. Editorial for special issue: Enzyme immobilization and its applications. Molecules 2019, 24, 4619. [Google Scholar] [CrossRef] [Green Version]
- Reis, C.L.B.; de Sousa, E.Y.A.; de Serpa, J.F.; Oliveira, R.C.; dos Santos, J.C.S. Design of immobilized enzyme biocatalysts: Drawbacks and opportunities. Quim. Nova 2019, 42, 768–783. [Google Scholar]
- Thompson, M.P.; Peñafiel, I.; Cosgrove, S.C.; Turner, N.J. Biocatalysis using immobilized enzymes in continuous flow for the synthesis of fine chemicals. Org. Process Res. Dev. 2019, 23, 9–18. [Google Scholar] [CrossRef]
- Grunwald, P. Immobilized biocatalysts. Catalysts 2018, 8, 386. [Google Scholar] [CrossRef] [Green Version]
- Schmidt-Dannert, S.; Zhang, G.; Johnston, T.; Quin, M.B.; Schmidt-Dannert, C. Building a toolbox of protein scaffolds for future immobilization of biocatalysts. Appl. Microbiol. Biotechnol. 2018, 102, 8373–8388. [Google Scholar] [CrossRef]
- Ren, S.; Li, C.; Jiao, X.; Jia, S.; Jiang, Y.; Bilal, M.; Cui, J. Recent progress in multienzymes co-immobilization and multienzyme system applications. Chem. Eng. J. 2019, 373, 1254–1278. [Google Scholar] [CrossRef]
- Sheldon, R.A. CLEAs, combi-CLEASs and ‘smart’ magnetic CLEAs: Biocatalysis in a bio-based economy. Catalysts 2019, 9, 261. [Google Scholar] [CrossRef] [Green Version]
- Priyanka, P.; Tan, Y.; Kinsella, G.K.; Henehan, G.T.; Ryan, B.J. Solvent stable microbial lipases: Current understanding and biotechnological applications. Biotechnol. Lett. 2019, 41, 203–220. [Google Scholar] [CrossRef] [Green Version]
- Filho, D.G.; Silva, A.G.; Guidini, C.Z. Lipases: Sources, immobilization methods, and industrial applications. Appl. Microbiol. Biotechnol. 2019, 103, 7399–7423. [Google Scholar] [CrossRef]
- Fernandez-Lorente, G.; Godoy, C.A.; Mendes, A.A.; Lopez-Gallego, F.; Grazu, V.; de las Rivas, B.; Palomo, J.M.; Hermoso, J.; Fernandez-Lafuente, R.; Guisan, J.M. Solid-phase chemical amination of a lipase from Bacillus thermocatenulatus to improve its stabilization via covalent immobilization on highly activated glyoxyl-agarose. Biomacromolecules 2008, 9, 2553–2561. [Google Scholar] [CrossRef]
- de Godoy Daiha, K.; Angeli, R.; de Oliveira, S.D.; Almeida, R.V. Are lipases still important biocatalysts? A study of scientific publications and patents for technological forecasting. PLoS ONE 2015, 10, e0131624. [Google Scholar]
- Dwivedee, B.P.; Soni, S.; Sharma, M.; Bhaumik, J.; Laha, J.K.; Banerjee, U.C. Promiscuity of lipase-catalyzed reactions for organic synthesis: A recent update. ChemistrySelect 2018, 3, 2441–2466. [Google Scholar] [CrossRef]
- Monteiro, R.R.C.; Virgen-Ortiz, J.J.; Berenguer-Murcia, Á.; da Rocha, T.N.; dos Santos, J.C.S.; Alcántara, A.R.; Fernandez-Lafuente, R. Biotechnological relevance of the lipase A from Candida antarctica. Catal. Today 2020. In press. [Google Scholar] [CrossRef]
- Thangaraj, B.; Solomon, P.R. Immobilization of lipases—A review. Part II: Carrier materials. ChemBioEng Rev. 2019, 6, 167–194. [Google Scholar] [CrossRef]
- Thangaraj, B.; Solomon, P.R. Immobilization of lipases—A review. Part I: Enzyme immobilization. ChemBioEng Rev. 2019, 6, 157–166. [Google Scholar] [CrossRef]
- Facin, B.R.; Melchiors, M.S.; Valério, A.; Oliveira, J.V.; Oliveira, D. de Driving Immobilized lipases as biocatalysts: 10 years state of the art and future prospects. Ind. Eng. Chem. Res. 2019, 58, 5358–5378. [Google Scholar] [CrossRef]
- del Monte-Martínez, A.; Cutiño-Avila, B.V.; González-Bacerio, J. Rational design strategy as a novel immobilization methodology applied to lipases and phospholipases. In Lipases and Phospholipases. Methods in Molecular Biology; Sandoval, G., Ed.; Humana Press: New York, NY, USA, 2018; pp. 243–283. ISBN 978-1-4939-8672-9. [Google Scholar]
- Rodrigues, R.C.; Virgen-Ortíz, J.J.; dos Santos, J.C.S.; Berenguer-Murcia, Á.; Alcantara, A.R.; Barbosa, O.; Ortiz, C.; Fernandez-Lafuente, R. Immobilization of lipases on hydrophobic supports: Immobilization mechanism, advantages, problems, and solutions. Biotechnol. Adv. 2019, 37, 746–770. [Google Scholar] [CrossRef] [Green Version]
- Matsumoto, T.; Yamada, R.; Ogino, H. Chemical treatments for modification and immobilization to improve the solvent-stability of lipase. World J. Microbiol. Biotechnol. 2019, 35, 193. [Google Scholar] [CrossRef]
- Rodrigues, R.C.; Berenguer-Murcia, Á.; Fernandez-Lafuente, R. Coupling chemical modification and immobilization to improve the catalytic performance of enzymes. Adv. Synth. Catal. 2011, 353, 2216–2238. [Google Scholar] [CrossRef]
- Hernandez, K.; Fernandez-Lafuente, R. Control of protein immobilization: Coupling immobilization and site-directed mutagenesis to improve biocatalyst or biosensor performance. Enzyme Microb. Technol. 2011, 48, 107–122. [Google Scholar] [CrossRef]
- Rueda, N.; dos Santos, J.C.S.; Ortiz, C.; Torres, R.; Barbosa, O.; Rodrigues, R.C.; Berenguer-Murcia, Á.; Fernandez-Lafuente, R. Chemical modification in the design of immobilized enzyme biocatalysts: Drawbacks and opportunities. Chem. Rec. 2016, 16, 1436–1455. [Google Scholar] [CrossRef]
- Rodrigues, R.C.; Barbosa, O.; Ortiz, C.; Berenguer-Murcia, Á.; Torres, R.; Fernandez-Lafuente, R. Amination of enzymes to improve biocatalyst performance: Coupling genetic modification and physicochemical tools. RSC Adv. 2014, 4, 38350–38374. [Google Scholar] [CrossRef] [Green Version]
- Fernandez-Lafuente, R. Stabilization of multimeric enzymes: Strategies to prevent subunit dissociation. Enzyme Microb. Technol. 2009, 45, 405–418. [Google Scholar] [CrossRef]
- Santiago, G.; Martínez-Martínez, M.; Alonso, S.; Bargiela, R.; Coscolín, C.; Golyshin, P.N.; Guallar, V.; Ferrer, M. Rational engineering of multiple active sites in an ester hydrolase. Biochemistry 2018, 57, 2245–2255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alonso, S.; Santiago, G.; Cea-Rama, I.; Fernandez-Lopez, L.; Coscolín, C.; Modregger, J.; Ressmann, A.K.; Martínez-Martínez, M.; Marrero, H.; Bargiela, R.; et al. Genetically engineered proteins with two active sites for enhanced biocatalysis and synergistic chemo- and biocatalysis. Nat. Catal. 2020, 3, 319–328. [Google Scholar] [CrossRef]
- Quail, M.A.; Otto, T.D.; Gu, Y.; Harris, S.R.; Skelly, T.F.; McQuillan, J.A.; Swerdlow, H.P.; Oyola, S.O. Optimal enzymes for amplifying sequencing libraries. Nat. Methods 2012, 9, 10–11. [Google Scholar] [CrossRef]
- Struvay, C.; Feller, G. Optimization to low temperature activity in psychrophilic enzymes. Int. J. Mol. Sci. 2012, 13, 11643–11665. [Google Scholar] [CrossRef] [Green Version]
- Turner, B.L. Variation in ph optima of hydrolytic enzyme activities in tropical rain forest soils. Appl. Environ. Microbiol. 2010, 76, 6485–6493. [Google Scholar] [CrossRef] [Green Version]
- de Oliveira Carvalho, J.; Orlanda, J.F.F. Heat stability and effect of pH on enzyme activity of polyphenol oxidase in buriti (Mauritia flexuosa Linnaeus f.) fruit extract. Food Chem. 2017, 233, 159–163. [Google Scholar] [CrossRef]
- Herlet, J.; Kornberger, P.; Roessler, B.; Glanz, J.; Schwarz, W.H.; Liebl, W.; Zverlov, V. V A new method to evaluate temperature vs. pH activity profiles for biotechnological relevant enzymes. Biotechnol. Biofuels 2017, 10, 234. [Google Scholar] [CrossRef] [Green Version]
- Krogdahl, Å.; Sundby, A.; Holm, H. Characteristics of digestive processes in Atlantic salmon (Salmo salar). Enzyme pH optima, chyme pH, and enzyme activities. Aquaculture 2015, 449, 27–36. [Google Scholar] [CrossRef]
- Prieto, M.A.; Vazquez, J.A.; Murado, M.A. A new and general model to describe, characterize, quantify and classify the interactive effects of temperature and pH on the activity of enzymes. Analyst 2015, 140, 3587–3602. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.; Zhu, L.; Chang, Y.; Liu, X.; Liu, Z.; Sun, H.; Li, X.; Yu, H.; Shen, Z. Microenvironmental pH changes in immobilized cephalosporin C acylase during a proton-producing reaction and regulation by a two-stage catalytic process. Bioresour. Technol. 2017, 223, 157–165. [Google Scholar] [CrossRef] [PubMed]
- Volpato, G.; Rodrigues, R.; Fernandez-Lafuente, R. Use of enzymes in the production of semi-synthetic penicillins and cephalosporins: Drawbacks and perspectives. Curr. Med. Chem. 2010, 17, 3855–3873. [Google Scholar] [CrossRef] [PubMed]
- Boniello, C.; Mayr, T.; Klimant, I.; Koenig, B.; Riethorst, W.; Nidetzky, B. Intraparticle concentration gradients for substrate and acidic product in immobilized cephalosporin C amidase and their dependencies on carrier characteristics and reaction parameters. Biotechnol. Bioeng. 2010, 106, 528–540. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Lafuente, R.; Rosell, C.M.; Piatkowska, B.; Guisán, J.M. Synthesis of antibiotics (cephaloglycin) catalyzed by penicillin G acylase: Evaluation and optimization of different synthetic approaches. Enzyme Microb. Technol. 1996, 19, 9–14. [Google Scholar] [CrossRef]
- de Camargo, A.C.; Regitano-d’Arce, M.A.B.; Biasoto, A.C.T.; Shahidi, F. Enzyme-assisted extraction of phenolics from winemaking by-products: Antioxidant potential and inhibition of alpha-glucosidase and lipase activities. Food Chem. 2016, 212, 395–402. [Google Scholar] [CrossRef] [Green Version]
- Bezerra, R.M.F.; Pinto, P.A.; Fraga, I.; Dias, A.A. Enzyme inhibition studies by integrated Michaelis–Menten equation considering simultaneous presence of two inhibitors when one of them is a reaction product. Comput. Methods Programs Biomed. 2016, 125, 2–7. [Google Scholar] [CrossRef]
- Cleland, W.W. The kinetics of enzyme-catalyzed reactions with two or more substrates or products: II. Inhibition: Nomenclature and theory. Biochim. Biophys. Acta Spec. Sect. Enzymol. Subj. 1963, 67, 173–187. [Google Scholar]
- Andrades, D.; Graebin, N.G.; Kadowaki, M.K.; Ayub, M.A.Z.; Fernandez-Lafuente, R.; Rodrigues, R.C. Immobilization and stabilization of different β-glucosidases using the glutaraldehyde chemistry: Optimal protocol depends on the enzyme. Int. J. Biol. Macromol. 2019, 129, 672–678. [Google Scholar] [CrossRef]
- de Andrades, D.; Graebin, N.G.; Ayub, M.A.Z.; Fernandez-Lafuente, R.; Rodrigues, R.C. Physico-chemical properties, kinetic parameters, and glucose inhibition of several beta-glucosidases for industrial applications. Process Biochem. 2019, 78, 82–90. [Google Scholar] [CrossRef]
- Avinash, V.S.; Panigrahi, P.; Suresh, C.G.; Pundle, A.V.; Ramasamy, S. Structural modelling of substrate binding and inhibition in penicillin V acylase from Pectobacterium atrosepticum. Biochem. Biophys. Res. Commun. 2013, 437, 538–543. [Google Scholar] [CrossRef] [PubMed]
- Pessela, B.C.C.; Mateo, C.; Fuentes, M.; Vian, A.; García, J.L.; Carrascosa, A.V.; Guisán, J.M.; Fernández-Lafuente, R. The immobilization of a thermophilic β-galactosidase on Sepabeads supports decreases product inhibition: Complete hydrolysis of lactose in dairy products. Enzyme Microb. Technol. 2003, 33, 199–205. [Google Scholar] [CrossRef]
- Cleland, W.W. The kinetics of enzyme-catalyzed reactions with two or more substrates or products: III. Prediction of initial velocity and inhibition patterns by inspection. Biochim. Biophys. Acta Spec. Sect. Enzymol. Subj. 1963, 67, 188–196. [Google Scholar]
- Mateo, C.; Monti, R.; Pessela, B.C.C.; Fuentes, M.; Torres, R.; Manuel Guisán, J.; Fernández-Lafuente, R. Immobilization of lactase from Kluyveromyces lactis greatly reduces the inhibition promoted by glucose. Full hydrolysis of lactose in milk. Biotechnol. Prog. 2004, 20, 1259–1262. [Google Scholar] [CrossRef] [PubMed]
- Crameri, A.; Raillard, S.-A.; Bermudez, E.; Stemmer, W.P.C. DNA shuffling of a family of genes from diverse species accelerates directed evolution. Nature 1998, 391, 288–291. [Google Scholar] [CrossRef] [PubMed]
- Romero, P.A.; Arnold, F.H. Exploring protein fitness landscapes by directed evolution. Nat. Rev. Mol. Cell Biol. 2009, 10, 866–876. [Google Scholar] [CrossRef] [Green Version]
- Cherry, J.R.; Fidantsef, A.L. Directed evolution of industrial enzymes: An update. Curr. Opin. Biotechnol. 2003, 14, 438–443. [Google Scholar] [CrossRef]
- Pereira, S.A.P.; Dyson, P.J.; Saraiva, M.L.M.F.S. Miniaturized technologies for high-throughput drug screening enzymatic assays and diagnostics—A review. TrAC Trends Anal. Chem. 2020, 126, 115862. [Google Scholar] [CrossRef]
- Shan, X.; Liu, L.; Zeng, W.; Chen, J.; Zhou, J. High throughput screening platform for a FAD-dependent L-sorbose sehydrogenase. Front. Bioeng. Biotechnol. 2020, 8, 194. [Google Scholar] [CrossRef] [PubMed]
- Fan, F.; Wood, K. Bioluminescent assays for high-throughput screening. Assay Drug Dev. Technol. 2007, 5, 127–136. [Google Scholar] [CrossRef] [Green Version]
- Inglese, J.; Auld, D.; Jadhav, A.; Johnson, R.; Simeonov, A.; Yasgar, A.; Zheng, W.; Austin, C. Quantitative high-throughput screening: A titration-based approach that efficiently identifies biological activities in large chemical libraries. Proc. Natl. Acad. Sci. USA. 2006, 103, 11473–11478. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drueckhammer, D.G.; Hennen, W.J.; Pederson, R.L.; Iii, C.F.B.; Gautheron, C.M.; Krach, T.; Wong, C.H. Enzyme catalysis in synthetic carbohydrate chemistry. Synthesis 1991, 1991, 499–525. [Google Scholar] [CrossRef]
- do Nascimento, M.A.; Gotardo, L.E.; Bastos, E.M.; Almeida, F.C.L.; Leão, R.A.C.; de Souza, R.O.M.A.; Wojcieszak, R.; Itabaiana, I. Regioselective acylation of levoglucosan catalyzed by Candida antarctica (CaLB) lipase immobilized on epoxy resin. Sustainability 2019, 11, 6044. [Google Scholar] [CrossRef] [Green Version]
- Riva, S.; Chopineau, J.; Kieboom, A.P.G.; Klibanov, A.M. Protease-catalyzed regioselective esterification of sugars and related compounds in anhydrous dimethylformamide. J. Am. Chem. Soc. 1988, 110, 584–589. [Google Scholar] [CrossRef]
- Therisod, M.; Klibanov, A.M. Regioselective acylation of secondary hydroxyl groups in sugars catalyzed by lipases in organic solvents. J. Am. Chem. Soc. 1987, 109, 3977–3981. [Google Scholar] [CrossRef]
- Ren, B.; Zhang, L.; Zhang, M. Progress on selective acylation of carbohydrate hydroxyl groups. As. J. Org. Chem. 2019, 8, 1813–1823. [Google Scholar] [CrossRef]
- De Ruysscher, D.; Pang, L.; De Graef, S.; Nautiyal, M.; De Borggraeve, W.M.; Rozenski, J.; Strelkov, S.V.; Weeks, S.D.; Van Aerschot, A. Acylated sulfonamide adenosines as potent inhibitors of the adenylate-forming enzyme superfamily. Eur. J. Med. Chem. 2019, 174, 252–264. [Google Scholar] [CrossRef]
- Liu, M.; Kong, J.-Q. The enzymatic biosynthesis of acylated steroidal glycosides and their cytotoxic activity. Acta Pharm. Sin. B 2018, 8, 981–994. [Google Scholar] [CrossRef]
- Yang, J.; Liu, Y.; Liang, X.; Yang, Y.; Li, Q. Enantio-, regio-, and chemoselective lipase-catalyzed polymer synthesis. Macromol. Biosci. 2018, 18, 1800131. [Google Scholar] [CrossRef]
- Du, L.; Jiang, Z.; Xu, L.; Zhou, N.; Shen, J.; Dong, Z.; Shen, L.; Wang, H.; Luo, X. Microfluidic reactor for lipase-catalyzed regioselective synthesis of neohesperidin ester derivatives and their antimicrobial activity research. Carbohydr. Res. 2018, 455, 32–38. [Google Scholar] [CrossRef]
- Li, X.; Xu, H.; Zhao, G.; Wu, H.; Yu, Y.; Lai, F.; Xiao, X. Highly efficient synthesis of arbutin esters catalyzed by whole cells of Candida parapsilosis. RSC Adv. 2018, 8, 10081–10088. [Google Scholar] [CrossRef] [Green Version]
- Inprakhon, P.; Wongthongdee, N.; Amornsakchai, T.; Pongtharankul, T.; Sunintaboon, P.; Wiemann, L.O.; Durand, A.; Sieber, V. Lipase-catalyzed synthesis of sucrose monoester: Increased productivity by combining enzyme pretreatment and non-aqueous biphasic medium. J. Biotechnol. 2017, 259, 182–190. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.F.; Lalonde, J.J.; Momongan, M.; Bergbreiter, D.E.; Wong, C.H. Lipase-catalyzed irreversible transesterifications using enol esters as acylating reagents: Preparative enantio- and regioselective syntheses of alcohols, glycerol derivatives, sugars and organometallics. J. Am. Chem. Soc. 1988, 110, 7200–7205. [Google Scholar] [CrossRef]
- Matsumura, S.; Maeda, S.; Yoshikawa, S. Molecular design of biodegradable poly(carboxylic acid). Polym. Mater. Sci. Eng. Proc. ACS Div. Polym. Mater. Sci. Eng. 1989, 60, 885. [Google Scholar]
- Corey, E.J. The mechanism of the decarboxylation of α,β- and β,γ-unsaturated malonic acid derivatives and the course of decarboxylative condensation reactions in pyridine. J. Am. Chem. Soc. 1952, 74, 5897–5905. [Google Scholar] [CrossRef]
- Albers, T.; Biagini, S.C.G.; Hibbs, D.E.; Hursthouse, M.B.; Malik, K.M.A.; North, M.; Uriarte, E.; Zagotto, G. Desymmetrisation of meso-anhydrides utilising (S)-proline derivatives. Synthesis 1996, 1996, 393–398. [Google Scholar] [CrossRef]
- Galston, A.W.; Sawhney, R.K. Polyamines in plant physiology. Plant Physiol. 1990, 94, 406–410. [Google Scholar] [CrossRef] [Green Version]
- Jänne, J.; Alhonen, L.; Leinonen, P. Polyamines: From molecular biology to clinical applications. Ann. Med. 1991, 23, 241–259. [Google Scholar] [CrossRef]
- Park, S.; Kazlauskas, R.J. Improved preparation and use of room-temperature ionic liquids in lipase-catalyzed enantio- and regioselective acylations. J. Org. Chem. 2001, 66, 8395–8401. [Google Scholar] [CrossRef]
- Kennedy, J.F.; Kumar, H.; Panesar, P.S.; Marwaha, S.S.; Goyal, R.; Parmar, A.; Kaur, S. Enzyme-catalyzed regioselective synthesis of sugar esters and related compounds. J. Chem. Technol. Biotechnol. 2006, 81, 866–876. [Google Scholar] [CrossRef]
- Nakamura, K.; Yamanaka, R.; Matsuda, T.; Harada, T. Recent developments in asymmetric reduction of ketones with biocatalysts. Tetrahedron. Asymmetry 2003, 14, 2659–2681. [Google Scholar] [CrossRef]
- Pellissier, H. Asymmetric domino reactions. Part B: Reactions based on the use of chiral catalysts and biocatalysts. Tetrahedron 2006, 62, 2143–2173. [Google Scholar] [CrossRef]
- Matsuda, T.; Yamanaka, R.; Nakamura, K. Recent progress in biocatalysis for asymmetric oxidation and reduction. Tetrahedron Asymmetry 2009, 20, 513–557. [Google Scholar] [CrossRef]
- Niwa, T.; Murayama, N.; Imagawa, Y.; Yamazaki, H. Regioselective hydroxylation of steroid hormones by human cytochromes P450. Drug Metab. Rev. 2015, 47, 89–110. [Google Scholar] [CrossRef] [PubMed]
- Schnepel, C.; Sewald, N. Enzymatic Halogenation: A Timely strategy for regioselective C−H activation. Chem. A Eur. J. 2017, 23, 12064–12086. [Google Scholar] [CrossRef]
- Chen, Z.; Zhao, J.; Jiang, S.; Wei, D. Recent research advancements on regioselective nitrilase: Fundamental and applicative aspects. Appl. Microbiol. Biotechnol. 2019, 103, 6393–6405. [Google Scholar] [CrossRef]
- Filice, M.; Molina, M.; Benaiges, M.D.; Abian, O.; Valero, F.; Palomo, J.M. Solid-surface activated recombinant Rhizopous oryzae lipase expressed in Pichia pastoris and chemically modified variants as efficient catalysts in the synthesis of hydroxy monodeprotected glycals. Catal. Sci. Technol. 2017, 7, 1766–1775. [Google Scholar] [CrossRef]
- Brabcová, J.; Blažek, J.; Krečmerová, M.; Vondrášek, J.; Palomo, J.; Zarevúcka, M. Regioselective palmitoylation of 9-(2,3-dihydroxy- propyl)adenine catalyzed by a glycopolymer-enzyme conjugate. Molecules 2016, 21, 648. [Google Scholar] [CrossRef] [Green Version]
- Vavříková, E.; Gavezzotti, P.; Purchartová, K.; Fuksová, K.; Biedermann, D.; Kuzma, M.; Riva, S.; Křen, V. Regioselective alcoholysis of silychristin acetates catalyzed by lipases. Int. J. Mol. Sci. 2015, 16, 11983–11995. [Google Scholar] [CrossRef] [Green Version]
- da Silva, M.R.; Montenegro, T.G.C.; de Mattos, M.C.; de Oliveira, M.d.C.F.; de Lemos, T.L.G.; de Gonzalo, G.; Lavandera, I.; Gotor-Fernández, V.; Gotor, V. Regioselective preparation of thiamphenicol esters through lipase-catalyzed processes. J. Braz. Chem. Soc. 2014, 25, 987–994. [Google Scholar]
- Cabrera, Z.; Palomo, J.M.; Fernandez-Lorente, G.; Fernandez-Lafuente, R.; Guisan, J.M. Partial and enantioselective hydrolysis of diethyl phenylmalonate by immobilized preparations of lipase from Thermomyces lanuginose. Enzyme Microb. Technol. 2007, 40, 1280–1285. [Google Scholar] [CrossRef]
- Cabrera, Z.; Lopez-Gallego, F.; Fernandez-Lorente, G.; Palomo, J.M.; Montes, T.; Grazu, V.; Guisán, J.M.; Fernández-Lafuente, R. Asymmetric hydrolysis of dimethyl phenylmalonate by immobilized penicillin G acylase from E. coli. Enzyme Microb. Technol. 2007, 40, 997–1000. [Google Scholar] [CrossRef]
- Jiang, Y.; Wang, C.; Ma, N.; Chen, J.; Liu, C.; Wang, F.; Xu, J.; Cong, Z. Regioselective aromatic O-demethylation with an artificial P450BM3 peroxygenase system. Catal. Sci. Technol. 2020, 10, 1219–1223. [Google Scholar] [CrossRef]
- Guarneri, A.; Westphal, A.H.; Leertouwer, J.; Lunsonga, J.; Franssen, M.C.R.; Opperman, D.J.; Hollmann, F.; van Berkel, W.J.H.; Paul, C.E. Flavoenzyme-mediated regioselective aromatic hydroxylation with coenzyme biomimetics. ChemCatChem 2020, 12, 1368–1375. [Google Scholar] [CrossRef] [Green Version]
- Hu, D.; Zong, X.-C.; Xue, F.; Li, C.; Hu, B.-C.; Wu, M.-C. Manipulating regioselectivity of an epoxide hydrolase for single enzymatic synthesis of (R)-1,2-diols from racemic epoxides. Chem. Commun. 2020, 56, 2799–2802. [Google Scholar] [CrossRef]
- Wan, N.; Zhou, X.; Ma, R.; Tian, J.; Wang, H.; Cui, B.; Han, W.; Chen, Y. Synthesis of chiral 5-aryl-2-oxazolidinones via halohydrin dehalogenase-catalyzed enantio- and regioselective ring-opening of styrene oxides. Adv. Synth. Catal. 2020, 362, 1201–1207. [Google Scholar] [CrossRef]
- Pereira, P.R.M.; de Araújo, J.O.; Silva, J.R.A.; Alves, C.N.; Lameira, J.; Lima, A.H. Exploring chloride selectivity and halogenase regioselectivity of the sall enzyme through quantum mechanical/molecular mechanical modeling. J. Chem. Inf. Model. 2020, 60, 738–746. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, K.; Garcia-Verdugo, E.; Porcar, R.; Fernandez-Lafuente, R. Hydrolysis of triacetin catalyzed by immobilized lipases: Effect of the immobilization protocol and experimental conditions on diacetin yield. Enzyme Microb. Technol. 2011, 48, 510–517. [Google Scholar] [CrossRef]
- Hirata, D.B.; Albuquerque, T.L.; Rueda, N.; Virgen-Ortíz, J.J.; Tacias-Pascacio, V.G.; Fernandez-Lafuente, R. Evaluation of different immobilized lipases in transesterification reactions using tributyrin: Advantages of the heterofunctional octyl agarose beads. J. Mol. Catal. B Enzym. 2016, 133, 117–123. [Google Scholar] [CrossRef]
- Hirata, D.B.; Albuquerque, T.L.; Rueda, N.; Sánchez-Montero, J.M.; Garcia-Verdugo, E.; Porcar, R.; Fernandez-Lafuente, R. Advantages of heterofunctional octyl supports: Production of 1,2-dibutyrin by specific and selective hydrolysis of tributyrin catalyzed by immobilized lipases. ChemistrySelect 2016, 1, 3259–3270. [Google Scholar] [CrossRef]
- Wang, J.; Liu, X.; Wang, X.-D.; Dong, T.; Zhao, X.-Y.; Zhu, D.; Mei, Y.-Y.; Wu, G.-H. Selective synthesis of human milk fat-style structured triglycerides from microalgal oil in a microfluidic reactor packed with immobilized lipase. Bioresour. Technol. 2016, 220, 132–141. [Google Scholar] [CrossRef] [PubMed]
- Eichmann, T.O.; Kumari, M.; Haas, J.T.; Farese, R.V.; Zimmermann, R.; Lass, A.; Zechner, R. Studies on the substrate and stereo/regioselectivity of adipose triglyceride lipase, hormone-sensitive lipase, and diacylglycerol-O-acyltransferases. J. Biol. Chem. 2012, 287, 41446–41457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rogalska, E.; Cudrey, C.; Ferrato, F.; Verger, R. Stereoselective hydrolysis of triglycerides by animal and microbial lipases. Chirality 1993, 5, 24–30. [Google Scholar] [CrossRef] [PubMed]
- Rogalska, E.; Ransac, S.; Verger, R. Stereoselectivity of lipases. II. Stereoselective hydrolysis of triglycerides by gastric and pancreatic lipases. J. Biol. Chem. 1990, 265, 20271–20276. [Google Scholar]
- Rodrigues, R.C.; Ayub, M.A.Z. Effects of the combined use of Thermomyces lanuginosus and lipases for the transesterification and hydrolysis of soybean oil. Process Biochem. 2011, 46, 682–688. [Google Scholar] [CrossRef] [Green Version]
- Lee, D.H.; Kim, J.M.; Shin, H.Y.; Kang, S.W.; Kim, S.W. Biodiesel production using a mixture of immobilized Rhizopus oryzae and Candida rugosa lipases. Biotechnol. Bioprocess Eng. 2006, 11, 522–525. [Google Scholar] [CrossRef]
- Tan, J.-N.; Dou, Y. Deep eutectic solvents for biocatalytic transformations: Focused lipase-catalyzed organic reactions. Appl. Microbiol. Biotechnol. 2020, 104, 1481–1496. [Google Scholar] [CrossRef]
- Bharathi, D.; Rajalakshmi, G. Microbial lipases: An overview of screening, production and purification. Biocatal. Agric. Biotechnol. 2019, 22, 101368. [Google Scholar] [CrossRef]
- Hasan, F.; Shah, A.A.; Hameed, A. Industrial applications of microbial lipases. Enzyme Microb. Technol. 2006, 39, 235–251. [Google Scholar] [CrossRef]
- Ching-Velasquez, J.; Fernández-Lafuente, R.; Rodrigues, R.C.; Plata, V.; Rosales-Quintero, A.; Torrestiana-Sánchez, B.; Tacias-Pascacio, V.G. Production and characterization of biodiesel from oil of fish waste by enzymatic catalysis. Renew. Energy 2020, 153, 1346–1354. [Google Scholar] [CrossRef]
- Mendoza-Ortiz, P.A.; Gama, R.S.; Gómez, O.C.; Luiz, J.H.H.; Fernandez-Lafuente, R.; Cren, E.C.; Mendes, A.A. Sustainable enzymatic synthesis of a solketal ester—Process optimization and evaluation of its antimicrobial activity. Catalysts 2020, 10, 218. [Google Scholar] [CrossRef] [Green Version]
- Zieniuk, B.; Fabiszewska, A.; Białecka-Florjańczyk, E. Screening of solvents for favoring hydrolytic activity of Candida antarctica lipase B. Bioprocess Biosyst. Eng. 2020, 43, 605–613. [Google Scholar] [CrossRef] [PubMed]
- Zou, X.; Nadege, K.; Ninette, I.; Wen, Y.; Wu, S.; Jiang, X.; Zhang, H.; Jin, Q.; Wang, X. Preparation of docosahexaenoic acid-rich diacylglycerol-rich oil by lipase-catalyzed glycerolysis of microbial oil from Schizochytrium sp. in a solvent-free system. J. Am. Oil Chem. Soc. 2020, 97, 263–270. [Google Scholar] [CrossRef]
- Bayout, I.; Bouzemi, N.; Guo, N.; Mao, X.; Serra, S.; Riva, S.; Secundo, F. Natural flavor ester synthesis catalyzed by lipases. Flavour Fragr. J. 2020, 35, 209–218. [Google Scholar] [CrossRef]
- Fernandes, K.V.; Cavalcanti, E.D.C.; Cipolatti, E.P.; Aguieiras, E.C.G.; Pinto, M.C.C.; Tavares, F.A.; da Silva, P.R.; Fernandez-Lafuente, R.; Arana-Peña, S.; Pinto, J.C.; et al. Enzymatic synthesis of biolubricants from by-product of soybean oil processing catalyzed by different biocatalysts of Candida rugosa lipase. Catal. Today 2020. In press. [Google Scholar] [CrossRef]
- Monteiro, R.R.C.; Neto, D.M.A.; Fechine, P.B.A.; Lopes, A.A.S.; Gonçalves, L.R.B.; dos Santos, J.C.S.; de Souza, M.C.M.; Fernandez-Lafuente, R. Ethyl butyrate synthesis catalyzed by lipases A and B from Candida antarctica immobilized onto magnetic nanoparticles. Improvement of biocatalysts’ performance under ultrasonic irradiation. Int. J. Mol. Sci. 2019, 20, 5807. [Google Scholar] [CrossRef] [Green Version]
- Barsé, L.Q.; Graebin, N.G.; Cipolatti, E.P.; Robert, J.M.; Pinto, M.C.C.; Pinto, J.C.C.S.; Freire, D.M.G.; Rodrigues, R.C. Production and optimization of isopropyl palmitate via biocatalytic route using home-made enzymatic catalysts. J. Chem. Technol. Biotechnol. 2019, 94, 389–397. [Google Scholar] [CrossRef]
- Fukuda, H.; Kondo, A.; Noda, H. Biodiesel fuel production by transesterification of oils. J. Biosci. Bioeng. 2001, 92, 405–416. [Google Scholar] [CrossRef]
- Xu, X. Production of specific-structured triacylglycerols by lipase-catalyzed reactions: A review. Eur. J. Lipid Sci. Technol. 2000, 102, 287–303. [Google Scholar] [CrossRef]
- Malcata, F.X.; Reyes, H.R.; Garcia, H.S.; Hill, C.G.; Amundson, C.H. Kinetics and mechanisms of reactions catalysed by immobilized lipases. Enzyme Microb. Technol. 1992, 14, 426–446. [Google Scholar] [CrossRef]
- Bornscheuer, U.T.; Bessler, C.; Srinivas, R.; Hari Krishna, S. Optimizing lipases and related enzymes for efficient application. Trends Biotechnol. 2002, 20, 433–437. [Google Scholar] [CrossRef]
- Abed, S.M.; Elbandy, M.; Abdel-Samie, M.A.; Ali, A.H.; Korma, S.A.; Noman, A.; Wei, W.; Jin, Q. Screening of lipases for production of novel structured lipids from single cell oils. Process Biochem. 2020, 91, 181–188. [Google Scholar] [CrossRef]
- Hult, K.; Berglund, P. Enzyme promiscuity: Mechanism and applications. Trends Biotechnol. 2007, 25, 231–238. [Google Scholar] [CrossRef] [PubMed]
- Kapoor, M.; Gupta, M.N. Lipase promiscuity and its biochemical applications. Process Biochem. 2012, 47, 555–569. [Google Scholar] [CrossRef]
- Brzozowski, A.M.; Derewenda, U.; Derewenda, Z.S.; Dodson, G.G.; Lawson, D.M.; Turkenburg, J.P.; Bjorkling, F.; Huge-Jensen, B.; Patkar, S.A.; Thim, L. A model for interfacial activation in lipases from the structure of a fungal lipase-inhibitor complex. Nature 1991, 351, 491–494. [Google Scholar] [CrossRef] [PubMed]
- Verger, R. ‘Interfacial activation’ of lipases: Facts and artifacts. Trends Biotechnol. 1997, 15, 32–38. [Google Scholar] [CrossRef]
- Khan, F.I.; Lan, D.; Durrani, R.; Huan, W.; Zhao, Z.; Wang, Y. The lid domain in lipases: Structural and functional determinant of enzymatic properties. Front. Bioeng. Biotechnol. 2017, 5, 16. [Google Scholar] [CrossRef] [Green Version]
- Kumar, A.; Dhar, K.; Kanwar, S.S.; Arora, P.K. Lipase catalysis in organic solvents: Advantages and applications. Biol. Proced. Online 2016, 18, 2. [Google Scholar] [CrossRef] [Green Version]
- Borrelli, G.M.; Trono, D. Recombinant lipases and phospholipases and their use as biocatalysts for industrial applications. Int. J. Mol. Sci. 2015, 16, 20774–20840. [Google Scholar] [CrossRef] [Green Version]
- Misra, N.N.; Pankaj, S.K.; Segat, A.; Ishikawa, K. Cold plasma interactions with enzymes in foods and model systems. Trends Food Sci. Technol. 2016, 55, 39–47. [Google Scholar] [CrossRef]
- Khan, N.R.; Rathod, V.K. Enzyme catalyzed synthesis of cosmetic esters and its intensification: A review. Process Biochem. 2015, 50, 1793–1806. [Google Scholar] [CrossRef]
- Carvalho, A.; Fonseca, T.; Mattos, M.; Oliveira, M.; Lemos, T.; Molinari, F.; Romano, D.; Serra, I. Recent advances in lipase-mediated preparation of pharmaceuticals and their intermediates. Int. J. Mol. Sci. 2015, 16, 29682–29716. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, B.H.; Akoh, C.C. Recent research trends on the enzymatic synthesis of structured lipids. J. Food Sci. 2015, 80, C1713–C1724. [Google Scholar] [CrossRef] [PubMed]
- Sarmah, N.; Revathi, D.; Sheelu, G.; Yamuna Rani, K.; Sridhar, S.; Mehtab, V.; Sumana, C. Recent advances on sources and industrial applications of lipases. Biotechnol. Prog. 2018, 34, 5–28. [Google Scholar] [CrossRef]
- Neta, N.S.; Teixeira, J.A.; Rodrigues, L.R. Sugar ester surfactants: Enzymatic synthesis and applications in food industry. Crit. Rev. Food Sci. Nutr. 2015, 55, 595–610. [Google Scholar] [CrossRef]
- Lopresto, C.G.; Naccarato, S.; Albo, L.; De Paola, M.G.; Chakraborty, S.; Curcio, S.; Calabrò, V. Enzymatic transesterification of waste vegetable oil to produce biodiesel. Ecotoxicol. Environ. Saf. 2015, 121, 229–235. [Google Scholar] [CrossRef]
- Craven, R.J.; Lencki, R.W. Symmetry, chirality and crystalline tendency: The polymorphism of triacylglycerols. Food Funct. 2012, 3, 228–233. [Google Scholar] [CrossRef]
- Hsu, F.-F.; Turk, J. Structural characterization of triacylglycerols as lithiated adducts by electrospray ionization mass spectrometry using low-energy collisionally activated dissociation on a triple stage quadrupole instrument. J. Am. Soc. Mass Spectrom. 1999, 10, 587–599. [Google Scholar] [CrossRef] [Green Version]
- Mu, H.; Porsgaard, T. The metabolism of structured triacylglycerols. Prog. Lipid Res. 2005, 44, 430–448. [Google Scholar] [CrossRef]
- Cheng, C.; Gross, M.L.; Pittenauer, E. Complete structural elucidation of triacylglycerols by tandem sector mass spectrometry. Anal. Chem. 1998, 70, 4417–4426. [Google Scholar] [CrossRef]
- Zarai, Z.; Eddehech, A.; Rigano, F.; Oteri, M.; Micalizzi, G.; Dugo, P.; Mondello, L.; Cacciola, F. Characterization of monoacylglycerols and diacylglycerols rich in polyunsaturated fatty acids produced by hydrolysis of Musteleus mustelus liver oil catalyzed by an immobilized bacterial lipase. J. Chromatogr. A 2020, 1613, 460692. [Google Scholar] [CrossRef] [PubMed]
- Monte Blanco, S.F.M.; Santos, J.S.; Feltes, M.M.C.; Dors, G.; Licodiedoff, S.; Lerin, L.A.; de Oliveira, D.; Ninow, J.L.; Furigo, A. Optimization of diacylglycerol production by glycerolysis of fish oil catalyzed by Lipozyme TL IM with Tween 65. Bioprocess Biosyst. Eng. 2015, 38, 2379–2388. [Google Scholar] [CrossRef] [PubMed]
- Xia, Q.; Akanbi, T.O.; Li, R.; Wang, B.; Yang, W.; Barrow, C.J. Lipase-catalysed synthesis of palm oil-omega-3 structured lipids. Food Funct. 2019, 10, 3142–3149. [Google Scholar] [CrossRef] [PubMed]
- Mota, D.A.; Rajan, D.; Heinzl, G.C.; Osório, N.M.; Gominho, J.; Krause, L.C.; Soares, C.M.F.; Nampoothiri, K.M.; Sukumaran, R.K.; Ferreira-Dias, S. Production of low-calorie structured lipids from spent coffee grounds or olive pomace crude oils catalyzed by immobilized lipase in magnetic nanoparticles. Bioresour. Technol. 2020, 307, 123223. [Google Scholar] [CrossRef]
- Moharana, T.R.; Rao, N.M. Substrate structure and computation guided engineering of a lipase for omega-3 fatty acid selectivity. PLoS ONE 2020, 15, e0231177. [Google Scholar] [CrossRef] [Green Version]
- Chen, M.; Gao, X.; Yang, W.; Sun, C.; Yang, J.; Zhang, H.; Song, Y. Discovery and characterization of a stable lipase with preference toward long-chain fatty acids. Biotechnol. Lett. 2020, 42, 171–180. [Google Scholar] [CrossRef]
- Cao, X.; Liao, L.; Feng, F. Purification and characterization of an extracellular lipase from Trichosporon sp. and its application in enrichment of omega-3 polyunsaturated fatty acids. LWT 2020, 118, 108692. [Google Scholar] [CrossRef]
- Saario, S.M.; Laitinen, J.T. Monoglyceride lipase as an enzyme hydrolyzing 2-arachidonoylglycerol. Chem. Biodivers. 2007, 4, 1903–1913. [Google Scholar] [CrossRef]
- Bornscheuer, U.T. Lipase-catalyzed syntheses of monoacylglycerols. Enzyme Microb. Technol. 1995, 17, 578–586. [Google Scholar] [CrossRef]
- Yuan, D.; Wu, Z.; Wang, Y. Evolution of the diacylglycerol lipases. Prog. Lipid Res. 2016, 64, 85–97. [Google Scholar] [CrossRef]
- Lee, W.J.; Zhang, Z.; Lai, O.M.; Tan, C.P.; Wang, Y. Diacylglycerol in food industry: Synthesis methods, functionalities, health benefits, potential risks and drawbacks. Trends Food Sci. Technol. 2020, 97, 114–125. [Google Scholar] [CrossRef]
- Wang, X.; Cheng, Y.; Jin, Q.; Wei, W.; Wang, X. Biocatalytic synthesis and characterization of sn-1/3 and sn-2 monoacylglycerols. Biotechnol. Lett. 2019, 41, 789–799. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Fruekilde, M.-B.; Xu, X. Suppression of acyl migration in enzymatic production of structured lipids through temperature programming. Food Chem. 2005, 92, 101–107. [Google Scholar] [CrossRef]
- Filice, M.; Bavaro, T.; Fernandez-Lafuente, R.; Pregnolato, M.; Guisan, J.M.; Palomo, J.M.; Terreni, M. Chemo-biocatalytic regioselective one-pot synthesis of different deprotected monosaccharides. Catal. Today 2009, 140, 11–18. [Google Scholar] [CrossRef]
- Li, W.; Du, W.; Li, Q.; Sun, T.; Liu, D. Study on acyl migration kinetics of partial glycerides: Dependence on temperature and water activity. J. Mol. Catal. B Enzym. 2010, 63, 17–22. [Google Scholar] [CrossRef]
- Cao, X.; Mangas-Sánchez, J.; Feng, F.; Adlercreutz, P. Acyl migration in enzymatic interesterification of triacylglycerols: Effects of lipases from Thermomyces lanuginosus and Rhizopus oryzae, support material, and water activity. Eur. J. Lipid Sci. Technol. 2016, 118, 1579–1587. [Google Scholar] [CrossRef]
- Peng, B.; Chen, F.; Liu, X.; Hu, J.-N.; Zheng, L.-F.; Li, J.; Deng, Z.-Y. Trace water activity could improve the formation of 1,3-oleic-2-medium chain-rich triacylglycerols by promoting acyl migration in the lipase RM IM catalyzed interesterification. Food Chem. 2020, 313, 126130. [Google Scholar] [CrossRef]
- Li, W.; Du, W.; Li, Q.; Li, R.; Liu, D. Dependence on the properties of organic solvent: Study on acyl migration kinetics of partial glycerides. Bioresour. Technol. 2010, 101, 5737–5742. [Google Scholar] [CrossRef]
- Mao, J.; Hu, Z.; Hu, J.; Zhu, X.; Xiong, H. A density functional theory (DFT) study of the acyl migration occurring during lipase-catalyzed transesterifications. Int. J. Mol. Sci. 2019, 20, 3438. [Google Scholar] [CrossRef] [Green Version]
- Oda, M.; Kaieda, M.; Hama, S.; Yamaji, H.; Kondo, A.; Izumoto, E.; Fukuda, H. Facilitatory effect of immobilized lipase-producing Rhizopus oryzae cells on acyl migration in biodiesel-fuel production. Biochem. Eng. J. 2005, 23, 45–51. [Google Scholar] [CrossRef]
- Xu, X.; Balchen, S.; Høy, C.-E.; Adler-Nissen, J. Pilot batch production of specific-structured lipids by lipase-catalyzed interesterification: Preliminary study on incorporation and acyl migration. J. Am. Oil Chem. Soc. 1998, 75, 301–308. [Google Scholar] [CrossRef]
- Briand, D.; Dubreucq, E.; Galzy, P. Functioning and regioselectivity of the lipase of Candida parapsilosis (Ashford) langeron and talice in aqueous medium: New interpretation of regioselectivity taking acyl migration into account. Eur. J. Biochem. 1995, 228, 169–175. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.K.-C.; Nozaki, K.; Wong, C.-H. Problems of acyl migration in lipase-catalyzed enantioselective transformation of meso-1,3-diol systems. Biocatal. Biotransformation 1990, 3, 169–177. [Google Scholar]
- Kim, B.H.; Akoh, C.C. Modeling of lipase-catalyzed acidolysis of sesame oil and caprylic acid by response surface methodology: Optimization of reaction conditions by considering both acyl incorporation and migration. J. Agric. Food Chem. 2005, 53, 8033–8037. [Google Scholar] [CrossRef]
- Kaieda, M.; Samukawa, T.; Matsumoto, T.; Ban, K.; Kondo, A.; Shimada, Y.; Noda, H.; Nomoto, F.; Ohtsuka, K.; Izumoto, E.; et al. Biodiesel fuel production from plant oil catalyzed by Rhizopus oryzae lipase in a water-containing system without an organic solvent. J. Biosci. Bioeng. 1999, 88, 627–631. [Google Scholar] [CrossRef]
- Li, W.; Li, R.; Li, Q.; Du, W.; Liu, D. Acyl migration and kinetics study of 1(3)-positional specific lipase of Rhizopus oryzae-catalyzed methanolysis of triglyceride for biodiesel production. Process Biochem. 2010, 45, 1888–1893. [Google Scholar] [CrossRef]
- Walia, S.; Mukhia, S.; Bhatt, V.; Kumar, R.; Kumar, R. Variability in chemical composition and antimicrobial activity of Tagetes minuta L. essential oil collected from different locations of Himalaya. Ind. Crops Prod. 2020, 150, 112449. [Google Scholar] [CrossRef]
- da Silva, C.M.; Zanqui, A.B.; Visentainer, J.V.; Cardozo-Filho, L.; Bittencourt, P.R.S.; Morais, D.R.; Santos, J.M.; Eberlin, M.N.; Matsushita, M. Quality and composition of three palm oils isolated by clean and sustainable process. J. Clean. Prod. 2020, 259, 120905. [Google Scholar] [CrossRef]
- Atabani, A.E.; Silitonga, A.S.; Ong, H.C.; Mahlia, T.M.I.; Masjuki, H.H.; Badruddin, I.A.; Fayaz, H. Non-edible vegetable oils: A critical evaluation of oil extraction, fatty acid compositions, biodiesel production, characteristics, engine performance and emissions production. Renew. Sustain. Energy Rev. 2013, 18, 211–245. [Google Scholar] [CrossRef]
- Hussain, A.I.; Anwar, F.; Hussain Sherazi, S.T.; Przybylski, R. Chemical composition, antioxidant and antimicrobial activities of basil (Ocimum basilicum) essential oils depends on seasonal variations. Food Chem. 2008, 108, 986–995. [Google Scholar] [CrossRef]
- Goswami, D.; Basu, J.K.; De, S. Lipase applications in oil hydrolysis with a case study on castor oil: A review. Crit. Rev. Biotechnol. 2013, 33, 81–96. [Google Scholar] [CrossRef] [PubMed]
- Shahinuzzaman, M.; Yaakob, Z.; Moniruzzaman, M. Medicinal and cosmetics soap production from Jatropha oil. J. Cosmet. Dermatol. 2016, 15, 185–193. [Google Scholar] [CrossRef] [PubMed]
- Murty, V.R.; Bhat, J.; Muniswaran, P.K.A. Hydrolysis of oils by using immobilized lipase enzyme: A review. Biotechnol. Bioprocess Eng. 2002, 7, 57–66. [Google Scholar] [CrossRef]
- Tan, T.; Lu, J.; Nie, K.; Deng, L.; Wang, F. Biodiesel production with immobilized lipase: A review. Biotechnol. Adv. 2010, 28, 628–634. [Google Scholar] [CrossRef] [PubMed]
- Bajaj, A.; Lohan, P.; Jha, P.N.; Mehrotra, R. Biodiesel production through lipase catalyzed transesterification: An overview. J. Mol. Catal. B Enzym. 2010, 62, 9–14. [Google Scholar] [CrossRef]
- Zhao, X.; Qi, F.; Yuan, C.; Du, W.; Liu, D. Lipase-catalyzed process for biodiesel production: Enzyme immobilization, process simulation and optimization. Renew. Sustain. Energy Rev. 2015, 44, 182–197. [Google Scholar] [CrossRef]
- Aguieiras, E.C.G.; Cavalcanti-Oliveira, E.D.; Freire, D.M.G. Current status and new developments of biodiesel production using fungal lipases. Fuel 2015, 159, 52–67. [Google Scholar] [CrossRef]
- Hama, S.; Noda, H.; Kondo, A. How lipase technology contributes to evolution of biodiesel production using multiple feedstocks. Curr. Opin. Biotechnol. 2018, 50, 57–64. [Google Scholar] [CrossRef]
- Zhong, L.; Feng, Y.; Wang, G.; Wang, Z.; Bilal, M.; Lv, H.; Jia, S.; Cui, J. Production and use of immobilized lipases in/on nanomaterials: A review from the waste to biodiesel production. Int. J. Biol. Macromol. 2020, 152, 207–222. [Google Scholar] [CrossRef]
- Wancura, J.H.C.; Tres, M.V.; Jahn, S.L.; Oliveira, J.V. Lipases in liquid formulation for biodiesel production: Current status and challenges. Biotechnol. Appl. Biochem. 2019. In press. [Google Scholar] [CrossRef]
- Cammarota, M.C.; Freire, D.M.G. A review on hydrolytic enzymes in the treatment of wastewater with high oil and grease content. Bioresour. Technol. 2006, 97, 2195–2210. [Google Scholar] [CrossRef] [PubMed]
- Rajendran, A.; Palanisamy, A.; Thangavelu, V. Lipase applications in food industry. Indian J. Biotechnol. 2007, 6, 141–158. [Google Scholar]
- Satyarthi, J.K.; Srinivas, D.; Ratnasamy, P. Hydrolysis of vegetable oils and fats to fatty acids over solid acid catalysts. Appl. Catal. A Gen. 2011, 391, 427–435. [Google Scholar] [CrossRef]
- Abdelmoez, W.; Mustafa, A. Oleochemical industry future through biotechnology. J. Oleo Sci. 2014, 63, 545–554. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdelmoez, W.; Mostafa, N.A.; Mustafa, A. Utilization of oleochemical industry residues as substrates for lipase production for enzymatic sunflower oil hydrolysis. J. Clean. Prod. 2013, 59, 290–297. [Google Scholar] [CrossRef]
- Edwinoliver, N.G.; Thirunavukarasu, K.; Purushothaman, S.; Rose, C.; Gowthaman, M.K.; Kamini, N.R. Corn steep liquor as a nutrition adjunct for the production of Aspergillus niger lipase and hydrolysis of oils thereof. J. Agric. Food Chem. 2009, 57, 10658–10663. [Google Scholar] [CrossRef] [PubMed]
- Alves, J.S.; Vieira, N.S.; Cunha, A.S.; Silva, A.M.; Záchia Ayub, M.A.; Fernandez-Lafuente, R.; Rodrigues, R.C. Combi-lipase for heterogeneous substrates: A new approach for hydrolysis of soybean oil using mixtures of biocatalysts. RSC Adv. 2014, 4, 6863–6868. [Google Scholar] [CrossRef] [Green Version]
- Shimada, Y.; Watanabe, Y.; Sugihara, A.; Tominaga, Y. Enzymatic alcoholysis for biodiesel fuel production and application of the reaction to oil processing. J. Mol. Catal. B Enzym. 2002, 17, 133–142. [Google Scholar] [CrossRef]
- Rodionova, M.V.; Poudyal, R.S.; Tiwari, I.; Voloshin, R.A.; Zharmukhamedov, S.K.; Nam, H.G.; Zayadan, B.K.; Bruce, B.D.; Hou, H.J.M.; Allakhverdiev, S.I. Biofuel production: Challenges and opportunities. Int. J. Hydrogen Energy 2017, 42, 8450–8461. [Google Scholar] [CrossRef]
- Živković, S.B.; Veljković, M.V.; Banković-Ilić, I.B.; Krstić, I.M.; Konstantinović, S.S.; Ilić, S.B.; Avramović, J.M.; Stamenković, O.S.; Veljković, V.B. Technological, technical, economic, environmental, social, human health risk, toxicological and policy considerations of biodiesel production and use. Renew. Sustain. Energy Rev. 2017, 79, 222–247. [Google Scholar] [CrossRef]
- Amini, Z.; Ilham, Z.; Ong, H.C.; Mazaheri, H.; Chen, W.-H. State of the art and prospective of lipase-catalyzed transesterification reaction for biodiesel production. Energy Convers. Manag. 2017, 141, 339–353. [Google Scholar] [CrossRef]
- Poppe, J.K.; Fernandez-Lafuente, R.; Rodrigues, R.C.; Ayub, M.A.Z. Enzymatic reactors for biodiesel synthesis: Present status and future prospects. Biotechnol. Adv. 2015, 33, 511–525. [Google Scholar] [CrossRef] [PubMed]
- Guldhe, A.; Singh, B.; Mutanda, T.; Permaul, K.; Bux, F. Advances in synthesis of biodiesel via enzyme catalysis: Novel and sustainable approaches. Renew. Sustain. Energy Rev. 2015, 41, 1447–1464. [Google Scholar] [CrossRef]
- Ribeiro, B.D.; de Castro, A.M.; Coelho, M.A.Z.; Freire, D.M.G. Production and use of lipases in bioenergy: A review from the feedstocks to biodiesel production. Enzyme Res. 2011, 2011, 615803. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ortiz, C.; Ferreira, M.L.; Barbosa, O.; Dos Santos, J.C.S.; Rodrigues, R.C.; Berenguer-Murcia, Á.; Briand, L.E.; Fernandez-Lafuente, R. Novozym 435: The “perfect” lipase immobilized biocatalyst? Catal. Sci. Technol. 2019, 9, 2380–2420. [Google Scholar] [CrossRef] [Green Version]
- Fernandez-Lafuente, R. Lipase from Thermomyces lanuginosus: Uses and prospects as an industrial biocatalyst. J. Mol. Catal. B Enzym. 2010, 62, 197–212. [Google Scholar] [CrossRef]
- Rodrigues, R.C.; Fernandez-Lafuente, R. Lipase from Rhizomucor miehei as an industrial biocatalyst in chemical process. J. Mol. Catal. B Enzym. 2010, 64, 1–22. [Google Scholar] [CrossRef]
- Rodrigues, R.C.; Fernandez-Lafuente, R. Lipase from Rhizomucor miehei as a biocatalyst in fats and oils modification. J. Mol. Catal. B Enzym. 2010, 66, 15–32. [Google Scholar] [CrossRef]
- Rodrigues, R.C.; Volpato, G.; Wada, K.; Ayub, M.A.Z. Enzymatic Synthesis of biodiesel from transesterification reactions of vegetable oils and short chain alcohols. J. Am. Oil Chem. Soc. 2008, 85, 925–930. [Google Scholar] [CrossRef]
- Ranganathan, S.V.; Narasimhan, S.L.; Muthukumar, K. An overview of enzymatic production of biodiesel. Bioresour. Technol. 2008, 99, 3975–3981. [Google Scholar] [CrossRef]
- Szczęsna Antczak, M.; Kubiak, A.; Antczak, T.; Bielecki, S. Enzymatic biodiesel synthesis—Key factors affecting efficiency of the process. Renew. Energy 2009, 34, 1185–1194. [Google Scholar] [CrossRef]
- Salis, A.; Pinna, M.; Monduzzi, M.; Solinas, V. Biodiesel production from triolein and short chain alcohols through biocatalysis. J. Biotechnol. 2005, 119, 291–299. [Google Scholar] [CrossRef] [PubMed]
- Aguieiras, E.C.G.; Cavalcanti-Oliveira, E.D.; de Castro, A.M.; Langone, M.A.P.; Freire, D.M.G. Biodiesel production from Acrocomia aculeata acid oil by (enzyme/enzyme) hydroesterification process: Use of vegetable lipase and fermented solid as low-cost biocatalysts. Fuel 2014, 135, 315–321. [Google Scholar] [CrossRef]
- de Sousa, J.S.; Cavalcanti-Oliveira, E.d.; Aranda, D.A.G.; Freire, D.M.G. Application of lipase from the physic nut (Jatropha curcas L.) to a new hybrid (enzyme/chemical) hydroesterification process for biodiesel production. J. Mol. Catal. B Enzym. 2010, 65, 133–137. [Google Scholar] [CrossRef]
- Soares, D.; Pinto, A.F.; Gonçalves, A.G.; Mitchell, D.A.; Krieger, N. Biodiesel production from soybean soapstock acid oil by hydrolysis in subcritical water followed by lipase-catalyzed esterification using a fermented solid in a packed-bed reactor. Biochem. Eng. J. 2013, 81, 15–23. [Google Scholar] [CrossRef]
- Bressani, A.P.P.; Garcia, K.C.A.; Hirata, D.B.; Mendes, A.A. Production of alkyl esters from macaw palm oil by a sequential hydrolysis/esterification process using heterogeneous biocatalysts: Optimization by response surface methodology. Bioprocess Biosyst. Eng. 2015, 38, 287–297. [Google Scholar] [CrossRef]
- Vescovi, V.; Rojas, M.J.; Baraldo, A.; Botta, D.C.; Santana, F.A.M.; Costa, J.P.; Machado, M.S.; Honda, V.K.; de Lima Camargo Giordano, R.; Tardioli, P.W. Lipase-catalyzed production of biodiesel by hydrolysis of waste cooking oil followed by esterification of free fatty acids. J. Am. Oil Chem. Soc. 2016, 93, 1615–1624. [Google Scholar] [CrossRef]
- Zenevicz, M.C.P.; Jacques, A.; de Oliveira, D.; Furigo, A.; Valério, A.; Oliveira, J.V. A two-step enzymatic strategy to produce ethyl esters using frying oil as substrate. Ind. Crops Prod. 2017, 108, 52–55. [Google Scholar] [CrossRef]
- Wancura, J.H.C.; Rosset, D.V.; Ugalde, G.A.; Oliveira, J.V.; Mazutti, M.A.; Tres, M.V.; Jahn, S.L. Feeding strategies of methanol and lipase on Eversa® Transform-mediated hydroesterification for FAME production. Can. J. Chem. Eng. 2019, 97, 1332–1339. [Google Scholar] [CrossRef]
- Rosset, D.V.; Wancura, J.H.C.; Ugalde, G.A.; Oliveira, J.V.; Tres, M.V.; Kuhn, R.C.; Jahn, S.L. Enzyme-catalyzed production of FAME by hydroesterification of soybean oil using the novel soluble lipase ns 40116. Appl. Biochem. Biotechnol. 2019, 188, 914–926. [Google Scholar] [CrossRef]
- Wancura, J.H.C.; Rosset, D.V.; Mazutti, M.A.; Ugalde, G.A.; de Oliveira, J.V.; Tres, M.V.; Jahn, S.L. Improving the soluble lipase–catalyzed biodiesel production through a two-step hydroesterification reaction system. Appl. Microbiol. Biotechnol. 2019, 103, 7805–7817. [Google Scholar] [CrossRef] [PubMed]
- Luque, R.; Clark, J.H. Biodiesel-like biofuels from simultaneous transesterification/esterification of waste oils with a biomass-derived solid acid catalyst. ChemCatChem 2011, 3, 594–597. [Google Scholar] [CrossRef]
- Calero, J.; Luna, D.; Sancho, E.D.; Luna, C.; Bautista, F.M.; Romero, A.A.; Posadillo, A.; Berbel, J.; Verdugo-Escamilla, C. An overview on glycerol-free processes for the production of renewable liquid biofuels, applicable in diesel engines. Renew. Sustain. Energy Rev. 2015, 42, 1437–1452. [Google Scholar] [CrossRef]
- Calero, J.; Verdugo, C.; Luna, D.; Sancho, E.D.; Luna, C.; Posadillo, A.; Bautista, F.M.; Romero, A.A. Selective ethanolysis of sunflower oil with Lipozyme RM IM, an immobilized Rhizomucor miehei lipase, to obtain a biodiesel-like biofuel, which avoids glycerol production through the monoglyceride formation. N. Biotechnol. 2014, 31, 596–601. [Google Scholar] [CrossRef]
- Luna, C.; Verdugo, C.; Sancho, E.; Luna, D.; Calero, J.; Posadillo, A.; Bautista, F.; Romero, A. Biocatalytic behaviour of immobilized Rhizopus oryzae lipase in the 1,3-selective ethanolysis of sunflower oil to obtain a biofuel similar to biodiesel. Molecules 2014, 19, 11419–11439. [Google Scholar] [CrossRef] [Green Version]
- Luna, C.; Verdugo, C.; Sancho, E.D.; Luna, D.; Calero, J.; Posadillo, A.; Bautista, F.M.; Romero, A.A. Production of a biodiesel-like biofuel without glycerol generation, by using Novozym 435, an immobilized Candida antarctica lipase. Bioresour. Bioprocess. 2014, 1, 11. [Google Scholar] [CrossRef] [Green Version]
- Verdugo, C.; Luna, D.; Posadillo, A.; Sancho, E.D.; Rodríguez, S.; Bautista, F.; Luque, R.; Marinas, J.M.; Romero, A.A. Production of a new second generation biodiesel with a low cost lipase derived from Thermomyces lanuginosus: Optimization by response surface methodology. Catal. Today 2011, 167, 107–112. [Google Scholar] [CrossRef]
- Poppe, J.K.; Matte, C.R.; do Peralba, M.C.R.; Fernandez-Lafuente, R.; Rodrigues, R.C.; Ayub, M.A.Z. Optimization of ethyl ester production from olive and palm oils using mixtures of immobilized lipases. Appl. Catal. A Gen. 2015, 490, 50–56. [Google Scholar] [CrossRef]
- Brocca, S.; Schmidt-Dannert, C.; Schmid, R.D.; Lotti, M.; Alberghina, L. Design, total synthesis, and functional overexpression of the Candida rugosa lipl gene coding for a major industrial lipase. Protein Sci. 1998, 7, 1415–1422. [Google Scholar] [CrossRef]
- Brocca, S.; Secundo, F.; Ossola, M.; Alberghina, L.; Carrea, G.; Lotti, M. Sequence of the lid affects activity and specificity of Candida rugosa lipase isoenzymes. Protein Sci. 2003, 12, 2312–2319. [Google Scholar] [CrossRef] [Green Version]
- Piamtongkam, R.; Duquesne, S.; Bordes, F.; Barbe, S.; André, I.; Marty, A.; Chulalaksananukul, W. Enantioselectivity of Candida rugosa lipases (Lip1, Lip3, and Lip4) towards 2-bromo phenylacetic acid octyl esters controlled by a single amino acid. Biotechnol. Bioeng. 2011, 108, 1749–1756. [Google Scholar] [CrossRef] [PubMed]
- Bertolini, M.C.; Laramée, L.; Thomas, D.Y.; Cygler, M.; Schrag, J.D.; Vernet, T. Polymorphism in the lipase genes of Geotrichum candidum strains. Eur. J. Biochem. 1994, 219, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Bertolini, M.C.; Schrag, J.D.; Cygler, M.; Ziomek, E.; Thomas, D.Y.; Vernet, T. Expression and characterization of Geotrichum candidum lipase I gene: Comparison of specificity profile with lipase II. Eur. J. Biochem. 1995, 228, 863–869. [Google Scholar] [CrossRef] [PubMed]
- van Kampen, M.D.; Rosenstein, R.; Götz, F.; Egmond, M.R. Cloning, purification and characterisation of Staphylococcus warneri lipase 2. Biochim. Biophys. Acta Protein Struct. Mol. Enzymol. 2001, 1544, 229–241. [Google Scholar] [CrossRef]
- Cunha, A.G.; Fernández-Lorente, G.; Gutarra, M.L.E.; Bevilaqua, J.V.; Almeida, R.V.; Paiva, L.M.C.; Fernández-Lafuente, R.; Guisán, J.M.; Freire, D.M.G. Separation and immobilization of lipase from Penicillium simplicissimum by selective adsorption on hydrophobic supports. Appl. Biochem. Biotechnol. 2009, 156, 133–145. [Google Scholar] [CrossRef]
- Chen, H.-P.; Hsiao, K.-F.; Wu, S.-H.; Wang, K.-T. Regioselectivity enhancement by partial purification of lipase from Aspergillus niger. Biotechnol. Lett. 1995, 17, 305–308. [Google Scholar] [CrossRef]
- Mendes, A.A.; Oliveira, P.C.; de Castro, H.F. Properties and biotechnological applications of porcine pancreatic lipase. J. Mol. Catal. B Enzym. 2012, 78, 119–134. [Google Scholar] [CrossRef]
- Ferreira, M.M.; de Oliveira, G.F.; Basso, R.C.; Mendes, A.A.; Hirata, D.B. Optimization of free fatty acid production by enzymatic hydrolysis of vegetable oils using a non-commercial lipase from Geotrichum candidum. Bioprocess Biosyst. Eng. 2019, 42, 1647–1659. [Google Scholar] [CrossRef]
- Segura, R.L.; Palomo, J.M.; Mateo, C.; Cortes, A.; Terreni, M.; Fernández-Lafuente, R.; Guisan, J.M. Different properties of the lipases contained in porcine pancreatic lipase extracts as enantioselective biocatalysts. Biotechnol. Prog. 2004, 20, 825–829. [Google Scholar] [CrossRef]
- Palomo, J.M.; Fernández-Lorente, G.; Mateo, C.; Fuentes, M.; Guisan, J.M.; Fernández-Lafuente, R. Enzymatic production of (3S,4R)-(−)-4-(4′-fluorophenyl)-6-oxo-piperidin-3-carboxylic acid using a commercial preparation from Candida antarctica A: The role of a contaminant esterase. Tetrahedron: Asymmetry 2002, 13, 2653–2659. [Google Scholar] [CrossRef]
- Segura, R.L.; Betancor, L.; Palomo, J.M.; Hidalgo, A.; Fernández-Lorente, G.; Terreni, M.; Mateo, C.; Cortés, A.; Fernández-Lafuente, R.; Guisán, J.M. Purification and identification of different lipases contained in PPL commercial extracts: A minor contaminant is the main responsible of most esterasic activity. Enzyme Microb. Technol. 2006, 39, 817–823. [Google Scholar] [CrossRef]
- Arana-Peña, S.; Carballares, D.; Morellon-Sterling, S.; Berenguer-Murcia, A.; Alcantara, A.R.; Rodrigues, R.C.; Fernandez-Lafuente, R. Enzyme co-immobilization: Always the biocatalyst designers’ choice…or not? Biotechnol. Adv. Accepted.
- Buaban, B.; Inoue, H.; Yano, S.; Tanapongpipat, S.; Ruanglek, V.; Champreda, V.; Pichyangkura, R.; Rengpipat, S.; Eurwilaichitr, L. Bioethanol production from ball milled bagasse using an on-site produced fungal enzyme cocktail and xylose-fermenting Pichia stipitis. J. Biosci. Bioeng. 2010, 110, 18–25. [Google Scholar] [CrossRef]
- Passos, C.P.; Yilmaz, S.; Silva, C.M.; Coimbra, M.A. Enhancement of grape seed oil extraction using a cell wall degrading enzyme cocktail. Food Chem. 2009, 115, 48–53. [Google Scholar] [CrossRef]
- Sørensen, H.R.; Pedersen, S.; Jørgensen, C.T.; Meyer, A.S. Enzymatic hydrolysis of wheat arabinoxylan by a recombinant “minimal” enzyme cocktail containing β-xylosidase and novel endo-1,4-β-xylanase and α-l-arabinofuranosidase activities. Biotechnol. Prog. 2007, 23, 100–107. [Google Scholar] [CrossRef] [PubMed]
- Verma, D.; Kanagaraj, A.; Jin, S.; Singh, N.D.; Kolattukudy, P.E.; Daniell, H. Chloroplast-derived enzyme cocktails hydrolyse lignocellulosic biomass and release fermentable sugars. Plant Biotechnol. J. 2010, 8, 332–350. [Google Scholar] [CrossRef] [Green Version]
- Yamada, R.; Taniguchi, N.; Tanaka, T.; Ogino, C.; Fukuda, H.; Kondo, A. Cocktail δ-integration: A novel method to construct cellulolytic enzyme expression ratio-optimized yeast strains. Microb. Cell Fact. 2010, 9, 32. [Google Scholar] [CrossRef] [Green Version]
- Brabcová, J.; Demianová, Z.; Vondrášek, J.; Jágr, M.; Zarevúcka, M.; Palomo, J.M. Highly selective purification of three lipases from Geotrichum candidum 4013 and their characterization and biotechnological applications. J. Mol. Catal. B Enzym. 2013, 98, 62–72. [Google Scholar] [CrossRef]
- Fernández-Lorente, G.; Ortiz, C.; Segura, R.L.; Fernández-Lafuente, R.; Guisán, J.M.; Palomo, J.M. Purification of different lipases from Aspergillus niger by using a highly selective adsorption on hydrophobic supports. Biotechnol. Bioeng. 2005, 92, 773–779. [Google Scholar] [CrossRef]
- Volpato, G.; Filice, M.; Ayub, M.A.Z.; Guisan, J.M.; Palomo, J.M. Single-step purification of different lipases from Staphylococcus warneri. J. Chromatogr. A 2010, 1217, 473–478. [Google Scholar] [CrossRef]
- Hirata, D.; Fernández-Lafuente, R.; Basso1, R.; Mendes, A.; Tavano, O.; Badino, A.; Oliveira, L.; Esperança, M.; Moreira, N.; Castro, P. High lipase production from Geotrichum candidum in reduced time using cottonseed oil: Optimization, easy purification and specificity characterization. J. Chem. Eng. Res. Updat. 2017, 3, 60–69. [Google Scholar] [CrossRef]
- Guldhe, A.; Singh, P.; Ansari, F.A.; Singh, B.; Bux, F. Biodiesel synthesis from microalgal lipids using tungstated zirconia as a heterogeneous acid catalyst and its comparison with homogeneous acid and enzyme catalysts. Fuel 2017, 187, 180–188. [Google Scholar] [CrossRef]
- Ren, H.; Du, W.; Lv, L.; Liu, D. Study on free lipase-catalyzed ethanolysis for biodiesel preparation in an oil/water biphasic system. J. Am. Oil Chem. Soc. 2011, 88, 1551–1555. [Google Scholar] [CrossRef]
- Zhao, X.; El-Zahab, B.; Brosnahan, R.; Perry, J.; Wang, P. An organic soluble lipase for water-free synthesis of biodiesel. Appl. Biochem. Biotechnol. 2007, 143, 236–243. [Google Scholar] [CrossRef] [Green Version]
- Firdaus, M.Y.; Brask, J.; Nielsen, P.M.; Guo, Z.; Fedosov, S. Kinetic model of biodiesel production catalyzed by free liquid lipase from Thermomyces lanuginosus. J. Mol. Catal. B Enzym. 2016, 133, 55–64. [Google Scholar] [CrossRef]
- Cesarini, S.; Diaz, P.; Nielsen, P.M. Exploring a new, soluble lipase for FAMEs production in water-containing systems using crude soybean oil as a feedstock. Process Biochem. 2013, 48, 484–487. [Google Scholar] [CrossRef]
- Chen, X.; Du, W.; Liu, D. Effect of several factors on soluble lipase-mediated biodiesel preparation in the biphasic aqueous-oil systems. World J. Microbiol. Biotechnol. 2008, 24, 2097–2102. [Google Scholar] [CrossRef]
- Wancura, J.H.C.; Rosset, D.V.; Brondani, M.; Mazutti, M.A.; Oliveira, J.V.; Tres, M.V.; Jahn, S.L. Soluble lipase-catalyzed synthesis of methyl esters using a blend of edible and nonedible raw materials. Bioprocess Biosyst. Eng. 2018, 41, 1185–1193. [Google Scholar] [CrossRef]
- Cesarini, S.; Pastor, F.I.J.; Diaz, P. Improvement of P. aeruginosa 42A2 lipase preparations for FAMEs production, both in immobilized and soluble form. J. Mol. Catal. B Enzym. 2014, 99, 1–7. [Google Scholar] [CrossRef]
- Lam, M.K.; Lee, K.T.; Mohamed, A.R. Homogeneous, heterogeneous and enzymatic catalysis for transesterification of high free fatty acid oil (waste cooking oil) to biodiesel: A review. Biotechnol. Adv. 2010, 28, 500–518. [Google Scholar] [CrossRef]
- Remonatto, D.; Santin, C.M.T.; de Oliveira, D.; Di Luccio, M.; de Oliveira, J.V. FAME Production from waste oils through commercial soluble lipase Eversa ® catalysis. Ind. Biotechnol. 2016, 12, 254–262. [Google Scholar] [CrossRef]
- Adewale, P.; Vithanage, L.N.; Christopher, L. Optimization of enzyme-catalyzed biodiesel production from crude tall oil using Taguchi method. Energy Convers. Manag. 2017, 154, 81–91. [Google Scholar] [CrossRef]
- Andrade, T.A.; Errico, M.; Christensen, K.V. Evaluation of reaction mechanisms and kinetic parameters for the transesterification of castor oil by liquid enzymes. Ind. Eng. Chem. Res. 2017, 56, 9478–9488. [Google Scholar] [CrossRef] [Green Version]
- Séverac, E.; Galy, O.; Turon, F.; Pantel, C.A.; Condoret, J.-S.; Monsan, P.; Marty, A. Selection of CalB immobilization method to be used in continuous oil transesterification: Analysis of the economical impact. Enzyme Microb. Technol. 2011, 48, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Dossat, V.; Combes, D.; Marty, A. Continuous enzymatic transesterification of high oleic sunflower oil in a packed bed reactor: Influence of the glycerol production. Enzyme Microb. Technol. 1999, 25, 194–200. [Google Scholar] [CrossRef]
- Marty, A.; Dossat, V.; Condoret, J.-S. Continuous operation of lipase-catalyzed reactions in nonaqueous solvents: Influence of the production of hydrophilic compounds. Biotechnol. Bioeng. 1997, 56, 232–237. [Google Scholar] [CrossRef]
- Castillo, E.; Dossat, V.; Marty, A.; Condoret, J.S.; Combes, D. The role of silica gel in lipase-catalyzed esterification reactions of high-polar substrates. J. Am. Oil Chem. Soc. 1997, 74, 77–85. [Google Scholar] [CrossRef]
- Martins, A.B.; Friedrich, J.L.R.; Cavalheiro, J.C.; Garcia-Galan, C.; Barbosa, O.; Ayub, M.A.Z.; Fernandez-Lafuente, R.; Rodrigues, R.C. Improved production of butyl butyrate with lipase from Thermomyces lanuginosus immobilized on styrene–divinylbenzene beads. Bioresour. Technol. 2013, 134, 417–422. [Google Scholar] [CrossRef] [Green Version]
- Graebin, N.G.; Martins, A.B.; Lorenzoni, A.S.G.; Garcia-Galan, C.; Fernandez-Lafuente, R.; Ayub, M.A.Z.; Rodrigues, R.C. Immobilization of lipase B from Candida antarctica on porous styrene–divinylbenzene beads improves butyl acetate synthesis. Biotechnol. Prog. 2012, 28, 406–412. [Google Scholar] [CrossRef]
- Poppe, J.K.; Garcia-Galan, C.; Matte, C.R.; Fernandez-Lafuente, R.; Rodrigues, R.C.; Ayub, M.A.Z. Optimization of synthesis of fatty acid methyl esters catalyzed by lipase B from Candida antarctica immobilized on hydrophobic supports. J. Mol. Catal. B Enzym. 2013, 94, 51–56. [Google Scholar] [CrossRef]
- Martins, A.B.; Schein, M.F.; Friedrich, J.L.R.; Fernandez-Lafuente, R.; Ayub, M.A.Z.; Rodrigues, R.C. Ultrasound-assisted butyl acetate synthesis catalyzed by Novozym 435: Enhanced activity and operational stability. Ultrason. Sonochem. 2013, 20, 1155–1160. [Google Scholar] [CrossRef] [PubMed]
- Paludo, N.; Alves, J.S.; Altmann, C.; Ayub, M.A.Z.; Fernandez-Lafuente, R.; Rodrigues, R.C. The combined use of ultrasound and molecular sieves improves the synthesis of ethyl butyrate catalyzed by immobilized Thermomyces lanuginosus lipase. Ultrason. Sonochem. 2015, 22, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Fallavena, L.P.; Antunes, F.H.F.; Alves, J.S.; Paludo, N.; Ayub, M.A.Z.; Fernandez-Lafuente, R.; Rodrigues, R.C. Ultrasound technology and molecular sieves improve the thermodynamically controlled esterification of butyric acid mediated by immobilized lipase from Rhizomucor miehei. RSC Adv. 2014, 4, 8675–8681. [Google Scholar] [CrossRef] [Green Version]
- Akita, H.; Umezawa, I.; Sakurai, I.; Oishi, T. Structural characteristics of lipid-lipase aggregates for enantioselective hydrolysis in organic solvents. Chem. Pharm. Bull. 1993, 41, 12–15. [Google Scholar] [CrossRef] [Green Version]
- Shimizu, N.; Akita, H.; Kawamata, T. Enantioselective reaction of (.+-.)-1-Hydroxy-5-methyl-3-vinylcyclohex-2-ene and its acyl derivatives using lipid-lipase aggregates in organic solvent. Chem. Pharm. Bull. 1996, 44, 665–669. [Google Scholar] [CrossRef] [Green Version]
- Dordick, J.S. Enzymatic catalysis in monophasic organic solvents. Enzyme Microb. Technol. 1989, 11, 194–211. [Google Scholar] [CrossRef]
- Klibanov, A.M. Enzymatic catalysis in anhydrous organic solvents. Trends Biochem. Sci. 1989, 14, 141–144. [Google Scholar] [CrossRef]
- Virgen-Ortíz, J.J.; Tacias-Pascacio, V.G.; Hirata, D.B.; Torrestiana-Sanchez, B.; Rosales-Quintero, A.; Fernandez-Lafuente, R. Relevance of substrates and products on the desorption of lipases physically adsorbed on hydrophobic supports. Enzyme Microb. Technol. 2017, 96, 30–35. [Google Scholar] [CrossRef]
- Virgen-Ortíz, J.J.; Peirce, S.; Tacias-Pascacio, V.G.; Cortes-Corberan, V.; Marzocchella, A.; Russo, M.E.; Fernandez-Lafuente, R. Reuse of anion exchangers as supports for enzyme immobilization: Reinforcement of the enzyme-support multiinteraction after enzyme inactivation. Process Biochem. 2016, 51, 1391–1396. [Google Scholar] [CrossRef]
- Virgen-Ortíz, J.J.; Pedrero, S.G.; Fernandez-Lopez, L.; Lopez-Carrobles, N.; Gorines, B.C.; Otero, C.; Fernandez-Lafuente, R. Desorption of lipases immobilized on octyl-agarose beads and coated with ionic polymers after thermal inactivation. Stronger adsorption of polymers/unfolded protein composites. Molecules 2017, 22, 91. [Google Scholar] [CrossRef] [Green Version]
- Virgen-Ortíz, J.J.; dos Santos, J.C.S.; Berenguer-Murcia, Á.; Barbosa, O.; Rodrigues, R.C.; Fernandez-Lafuente, R. Polyethylenimine: A very useful ionic polymer in the design of immobilized enzyme biocatalysts. J. Mater. Chem. B 2017, 5, 7461–7490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tacias-Pascacio, V.G.; Ortiz, C.; Rueda, N.; Berenguer-Murcia, Á.; Acosta, N.; Aranaz, I.; Civera, C.; Fernandez-Lafuente, R.; Alcántara, A.R. Dextran aldehyde in biocatalysis: More than a mere immobilization system. Catalysts 2019, 9, 622. [Google Scholar] [CrossRef] [Green Version]
- Palomo, J.M.; Fuentes, M.; Fernández-Lorente, G.; Mateo, C.; Guisan, J.M.; Fernández-Lafuente, R. General trend of lipase to self-assemble giving bimolecular aggregates greatly modifies the enzyme functionality. Biomacromolecules 2003, 4, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Lorente, G.; Palomo, J.M.; Fuentes, M.; Mateo, C.; Guisán, J.M.; Fernández-Lafuente, R. Self-assembly of Pseudomonas fluorescens lipase into bimolecular aggregates dramatically affects functional properties. Biotechnol. Bioeng. 2003, 82, 232–237. [Google Scholar] [CrossRef]
- Palomo, J.M.; Ortiz, C.; Fuentes, M.; Fernandez-Lorente, G.; Guisan, J.M.; Fernandez-Lafuente, R. Use of immobilized lipases for lipase purification via specific lipase-lipase interactions. J. Chromatogr. A 2004, 1038, 267–273. [Google Scholar] [CrossRef]
- Palomo, J.M.; Ortiz, C.; Fernández-Lorente, G.; Fuentes, M.; Guisán, J.M.; Fernández-Lafuente, R. Lipase-lipase interactions as a new tool to immobilize and modulate the lipase properties. Enzyme Microb. Technol. 2005, 36, 447–454. [Google Scholar] [CrossRef]
- Palomo, J.M.; Peñas, M.M.; Fernández-Lorente, G.; Mateo, C.; Pisabarro, A.G.; Fernández-Lafuente, R.; Ramírez, L.; Guisán, J.M. Solid-phase handling of hydrophobins: Immobilized hydrophobins as a new tool to study lipases. Biomacromolecules 2003, 4, 204–210. [Google Scholar] [CrossRef]
- Park, Y.K.; Pastore, G.M.; de Almeida, M.M. Hydrolysis of soybean oil by a combined lipase system. J. Am. Oil Chem. Soc. 1988, 65, 252–254. [Google Scholar] [CrossRef]
- Qiao, H.; Zhang, F.; Guan, W.; Zuo, J.; Feng, D. Optimisation of combi-lipases from Aspergillus niger for the synergistic and efficient hydrolysis of soybean oil. Anim. Sci. J. 2017, 88, 772–780. [Google Scholar] [CrossRef]
- Huang, J.; Zhao, Q.; Bu, W.; Zhang, C.; Yang, Z.; Zhang, X.; Zhang, K. Ultrasound-assisted hydrolysis of lard for free fatty acids catalyzed by combined two lipases in aqueous medium. Bioengineered 2020, 11, 241–250. [Google Scholar] [CrossRef] [Green Version]
- Guan, F.; Peng, P.; Wang, G.; Yin, T.; Peng, Q.; Huang, J.; Guan, G.; Li, Y. Combination of two lipases more efficiently catalyzes methanolysis of soybean oil for biodiesel production in aqueous medium. Process Biochem. 2010, 45, 1677–1682. [Google Scholar] [CrossRef]
- Karmee, S.K. Preparation of biodiesel from nonedible oils using a mixture of used lipases. Energy Sources, Part A Recover. Util. Environ. Eff. 2016, 38, 2727–2733. [Google Scholar] [CrossRef]
- Zeng, L.; He, Y.; Jiao, L.; Li, K.; Yan, Y. Preparation of biodiesel with liquid synergetic lipases from rapeseed oil deodorizer distillate. Appl. Biochem. Biotechnol. 2017, 183, 778–791. [Google Scholar] [CrossRef] [PubMed]
- Amoah, J.; Ho, S.H.; Hama, S.; Yoshida, A.; Nakanishi, A.; Hasunuma, T.; Ogino, C.; Kondo, A. Conversion of Chlamydomonas sp. JSC4 lipids to biodiesel using Fusarium heterosporum lipase-expressing Aspergillus oryzae whole-cell as biocatalyst. Algal Res. 2017, 28, 16–23. [Google Scholar] [CrossRef]
- Jang, M.G.; Kim, D.K.; Park, S.C.; Lee, J.S.; Kim, S.W. Biodiesel production from crude canola oil by two-step enzymatic processes. Renew. Energy 2012, 42, 99–104. [Google Scholar] [CrossRef]
- Cesarini, S.; Haller, R.; Diaz, P.; Nielsen, P. Combining phospholipases and a liquid lipase for one-step biodiesel production using crude oils. Biotechnol. Biofuels 2014, 7, 29. [Google Scholar] [CrossRef] [Green Version]
- Amoah, J.; Ho, S.-H.; Hama, S.; Yoshida, A.; Nakanishi, A.; Hasunuma, T.; Ogino, C.; Kondo, A. Lipase cocktail for efficient conversion of oils containing phospholipids to biodiesel. Bioresour. Technol. 2016, 211, 224–230. [Google Scholar] [CrossRef] [Green Version]
- Hirose, T.; Yamauchi-Sato, Y.; Arai, Y.; Negishi, S. Synthesis of triacylglycerol containing conjugated linoleic acid by esterification using two blended lipases. J. Am. Oil Chem. Soc. 2006, 83, 35–38. [Google Scholar] [CrossRef]
- McNeill, G.P.; Yamane, T. Further improvements in the yield of monoglycerides during enzymatic glycerolysis of fats and oils. J. Am. Oil Chem. Soc. 1991, 68, 6–10. [Google Scholar] [CrossRef]
- Speranza, P.; Ribeiro, A.P.B.; Macedo, G.A. Lipase catalyzed interesterification of Amazonian patauá oil and palm stearin for preparation of specific-structured oils. J. Food Sci. Technol. 2015, 52, 8268–8275. [Google Scholar] [CrossRef] [Green Version]
- Macrae, A.R. Lipase-catalyzed interesterification of oils and fats. J. Am. Oil Chem. Soc. 1983, 60, 291–294. [Google Scholar] [CrossRef]
- Soumanou, M.M.; Pérignon, M.; Villeneuve, P. Lipase-catalyzed interesterification reactions for human milk fat substitutes production: A review. Eur. J. Lipid Sci. Technol. 2013, 115, 270–285. [Google Scholar] [CrossRef]
- Utama, Q.D.; Sitanggang, A.B.; Adawiyah, D.R.; Hariyadi, P. Lipase-catalyzed interesterification for the synthesis of medium-long-medium (MLM) structured lipids. Food Technol. Biotechnol. 2019, 57, 305–318. [Google Scholar] [CrossRef]
- Speranza, P.; Ribeiro, A.P.B.; Macedo, G.A. Application of lipases to regiospecific interesterification of exotic oils from an Amazonian area. J. Biotechnol. 2016, 218, 13–20. [Google Scholar] [CrossRef]
- Di Cosimo, R.; Mc Auliffe, J.; Poulose, A.J.; Bohlmann, G. Industrial use of immobilized enzymes. Chem. Soc. Rev. 2013, 42, 6437–6474. [Google Scholar] [CrossRef]
- Liese, A.; Hilterhaus, L. Evaluation of immobilized enzymes for industrial applications. Chem. Soc. Rev. 2013, 42, 6236–6249. [Google Scholar] [CrossRef]
- Iyer, P.V.; Ananthanarayan, L. Enzyme stability and stabilization—aqueous and non-aqueous environment. Process Biochem. 2008, 43, 1019–1032. [Google Scholar] [CrossRef]
- Mohamad, N.R.; Marzuki, N.H.C.; Buang, N.A.; Huyop, F.; Wahab, R.A. An overview of technologies for immobilization of enzymes and surface analysis techniques for immobilized enzymes. Biotechnol. Biotechnol. Equip. 2015, 29, 205–220. [Google Scholar] [CrossRef]
- Barbosa, O.; Ortiz, C.; Berenguer-Murcia, Á.; Torres, R.; Rodrigues, R.C.; Fernandez-Lafuente, R. Strategies for the one-step immobilization-purification of enzymes as industrial biocatalysts. Biotechnol. Adv. 2015, 33, 435–456. [Google Scholar] [CrossRef] [Green Version]
- Santos, J.C.S.D.; Barbosa, O.; Ortiz, C.; Berenguer-Murcia, A.; Rodrigues, R.C.; Fernandez-Lafuente, R. Importance of the support properties for immobilization or purification of enzymes. ChemCatChem 2015, 7, 2413–2432. [Google Scholar] [CrossRef] [Green Version]
- Guzik, U.; Hupert-Kocurek, K.; Wojcieszyńska, D. Immobilization as a strategy for improving enzyme properties- Application to oxidoreductases. Molecules 2014, 19, 8995–9018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, M.; Li, Q.; Zhao, X.; Du, W.; Liu, C.-M.; Liu, D.-H. Recovery of free lipase in the aqueous phase of enzymatic production of biodiesel by foam separation. Guocheng Gongcheng Xuebao/Chin. J. Process Eng. 2010, 10, 1187–1192. [Google Scholar]
- Gonçalves, M.C.P.; Kieckbusch, T.G.; Perna, R.F.; Fujimoto, J.T.; Morales, S.A.V.; Romanelli, J.P. Trends on enzyme immobilization researches based on bibliometric analysis. Process Biochem. 2019, 76, 95–110. [Google Scholar] [CrossRef]
- Barbosa, O.; Torres, R.; Ortiz, C.; Berenguer-Murcia, A.; Rodrigues, R.C.; Fernandez-Lafuente, R. Heterofunctional supports in enzyme immobilization: From traditional immobilization protocols to opportunities in tuning enzyme properties. Biomacromolecules 2013, 14, 2433–2462. [Google Scholar] [CrossRef] [Green Version]
- Manoel, E.A.; dos Santos, J.C.S.; Freire, D.M.G.; Rueda, N.; Fernandez-Lafuente, R. Immobilization of lipases on hydrophobic supports involves the open form of the enzyme. Enzyme Microb. Technol. 2015, 71, 53–57. [Google Scholar] [CrossRef]
- Kim, K.K.; Song, H.K.; Shin, D.H.; Hwang, K.Y.; Suh, S.W. The crystal structure of a triacylglycerol lipase from Pseudomonas cepacia reveals a highly open conformation in the absence of a bound inhibitor. Structure 1997, 5, 173–185. [Google Scholar] [CrossRef] [Green Version]
- Jaeger, K.-E.; Ransac, S.; Koch, H.B.; Ferrato, F.; Dijkstra, B.W. Topological characterization and modeling of the 3D structure of lipase from Pseudomonas aeruginosa. FEBS Lett. 1993, 332, 143–149. [Google Scholar] [CrossRef] [Green Version]
- Cygler, M.; Schrag, J.D. Structure and conformational flexibility of Candida rugosa lipase 11NRC. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1999, 1441, 205–214. [Google Scholar] [CrossRef]
- Dos Santos, J.C.S.; Rueda, N.; Torres, R.; Barbosa, O.; Gonçalves, L.R.B.; Fernandez-Lafuente, R. Evaluation of divinylsulfone activated agarose to immobilize lipases and to tune their catalytic properties. Process Biochem. 2015, 50, 918–927. [Google Scholar] [CrossRef]
- Dos Santos, J.C.S.; Rueda, N.; Sanchez, A.; Villalonga, R.; Gonçalves, L.R.B.; Fernandez-Lafuente, R. Versatility of divinylsulfone supports permits the tuning of CALB properties during its immobilization. RSC Adv. 2015, 5, 35801–35810. [Google Scholar] [CrossRef]
- Arana-Peña, S.; Rios, N.S.; Carballares, D.; Mendez-Sanchez, C.; Lokha, Y.; Gonçalves, L.R.B.; Fernandez-Lafuente, R. Effects of enzyme loading and immobilization conditions on the catalytic features of lipase from Pseudomonas fluorescens immobilized on octyl-agarose beads. Front. Bioeng. Biotechnol. 2020, 8, 36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lokha, Y.; Arana-Peña, S.; Rios, N.S.; Mendez-Sanchez, C.; Gonçalves, L.R.B.; Lopez-Gallego, F.; Fernandez-Lafuente, R. Modulating the properties of the lipase from Thermomyces lanuginosus immobilized on octyl agarose beads by altering the immobilization conditions. Enzyme Microb. Technol. 2020, 133, 109461. [Google Scholar] [CrossRef] [PubMed]
- Arana-Peña, S.; Rios, N.S.; Carballares, D.; Gonçalves, L.R.B.; Fernandez-Lafuente, R. Immobilization of lipases via interfacial activation on hydrophobic supports: Production of biocatalysts libraries by altering the immobilization conditions. Catal. Today 2020. In press. [Google Scholar] [CrossRef]
- Rueda, N.; dos Santos, J.C.S.; Torres, R.; Ortiz, C.; Barbosa, O.; Fernandez-Lafuente, R. Improved performance of lipases immobilized on heterofunctional octyl-glyoxyl agarose beads. RSC Adv. 2015, 5, 11212–11222. [Google Scholar] [CrossRef] [Green Version]
- Albuquerque, T.L.D.; Rueda, N.; Dos Santos, J.C.S.; Barbosa, O.; Ortiz, C.; Binay, B.; Özdemir, E.; Gonçalves, L.R.B.; Fernandez-Lafuente, R. Easy stabilization of interfacially activated lipases using heterofunctional divinyl sulfone activated-octyl agarose beads. Modulation of the immobilized enzymes by altering their nanoenvironment. Process Biochem. 2016, 51, 865–874. [Google Scholar] [CrossRef]
- Suescun, A.; Rueda, N.; dos Santos, J.C.S.; Castillo, J.J.; Ortiz, C.; Torres, R.; Barbosa, O.; Fernandez-Lafuente, R. Immobilization of lipases on glyoxyl–octyl supports: Improved stability and reactivation strategies. Process Biochem. 2015, 50, 1211–1217. [Google Scholar] [CrossRef]
- Rueda, N.; Dos Santos, C.S.; Rodriguez, M.D.; Albuquerque, T.L.; Barbosa, O.; Torres, R.; Ortiz, C.; Fernandez-Lafuente, R. Reversible immobilization of lipases on octyl-glutamic agarose beads: A mixed adsorption that reinforces enzyme immobilization. J. Mol. Catal. B Enzym. 2016, 128, 10–18. [Google Scholar] [CrossRef]
- Rueda, N.; dos Santos, J.C.S.; Ortiz, C.; Barbosa, O.; Fernandez-Lafuente, R.; Torres, R. Chemical amination of lipases improves their immobilization on octyl-glyoxyl agarose beads. Catal. Today 2016, 259, 107–118. [Google Scholar] [CrossRef]
- Rueda, N.; Albuquerque, T.; Bartolome-Cabrero, R.; Fernandez-Lopez, L.; Torres, R.; Ortiz, C.; dos Santos, J.; Barbosa, O.; Fernandez-Lafuente, R. Reversible immobilization of lipases on heterofunctional octyl-amino agarose beads prevents enzyme desorption. Molecules 2016, 21, 646. [Google Scholar] [CrossRef] [Green Version]
- Fernandez-Lopez, L.; Rueda, N.; Bartolome-Cabrero, R.; Rodriguez, M.D.; Albuquerque, T.L.; Dos Santos, J.C.S.; Barbosa, O.; Fernandez-Lafuente, R. Improved immobilization and stabilization of lipase from Rhizomucor miehei on octyl-glyoxyl agarose beads by using CaCl2. Process Biochem. 2016, 51, 48–52. [Google Scholar] [CrossRef]
- Rueda, N.; Dos Santos, J.C.S.; Torres, R.; Barbosa, O.; Ortiz, C.; Fernandez-Lafuente, R. Reactivation of lipases by the unfolding and refolding of covalently immobilized biocatalysts. RSC Adv. 2015, 5, 55588–55594. [Google Scholar] [CrossRef] [Green Version]
- Rios, N.S.; Mendez-Sanchez, C.; Arana-Peña, S.; Rueda, N.; Ortiz, C.; Gonçalves, L.R.B.; Fernandez-Lafuente, R. Immobilization of lipase from Pseudomonas fluorescens on glyoxyl-octyl-agarose beads: Improved stability and reusability. Biochim. Biophys. Acta Proteins Proteom. 2019, 1867, 741–747. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Lopez, L.; Pedrero, S.G.; Lopez-Carrobles, N.; Virgen-Ortíz, J.J.; Gorines, B.C.; Otero, C.; Fernandez-Lafuente, R. Physical crosslinking of lipase from Rhizomucor miehei immobilized on octyl agarose via coating with ionic polymers. Process Biochem. 2017, 54, 81–88. [Google Scholar] [CrossRef]
- Zaak, H.; Fernandez-Lopez, L.; Otero, C.; Sassi, M.; Fernandez-Lafuente, R. Improved stability of immobilized lipases via modification with polyethylenimine and glutaraldehyde. Enzyme Microb. Technol. 2017, 106, 67–74. [Google Scholar] [CrossRef]
- Fernandez-Lopez, L.; Virgen-OrtÍz, J.J.; Pedrero, S.G.; Lopez-Carrobles, N.; Gorines, B.C.; Otero, C.; Fernandez-Lafuente, R. Optimization of the coating of octyl-CALB with ionic polymers to improve stability and decrease enzyme leakage. Biocatal. Biotransform. 2018, 36, 47–56. [Google Scholar] [CrossRef]
- Lee, J.H.; Lee, D.H.; Lim, J.S.; Um, B.H.; Park, C.; Kang, S.W.; Kim, S.W. Optimization of the process for biodiesel production using a mixture of immobilized Rhizopus oryzae and Candida rugosa lipases. J. Microbiol. Biotechnol. 2008, 18, 1927–1931. [Google Scholar]
- Lee, J.H.; Kim, S.B.; Park, C.; Tae, B.; Han, S.O.; Kim, S.W. Development of batch and continuous processes on biodiesel production in a packed-bed reactor by a mixture of immobilized Candida rugosa and Rhizopus oryzae lipases. Appl. Biochem. Biotechnol. 2010, 161, 365–371. [Google Scholar] [CrossRef]
- Lee, J.H.; Kim, S.B.; Kang, S.W.; Song, Y.S.; Park, C.; Han, S.O.; Kim, S.W. Biodiesel production by a mixture of Candida rugosa and Rhizopus oryzae lipases using a supercritical carbon dioxide process. Bioresour. Technol. 2011, 102, 2105–2108. [Google Scholar] [CrossRef]
- Li, Q.; Zheng, J.; Yan, Y. Biodiesel preparation catalyzed by compound-lipase in co-solvent. Fuel Process. Technol. 2010, 91, 1229–1234. [Google Scholar] [CrossRef]
- Huang, Y.; Zheng, H.; Yan, Y. Optimization of lipase-catalyzed transesterification of lard for biodiesel production using response surface methodology. Appl. Biochem. Biotechnol. 2010, 160, 504–515. [Google Scholar] [CrossRef]
- Liu, Y.; Yan, Y.; Hu, F.; Yao, A.; Wang, Z.; Wei, F. Transesterification for biodiesel production catalyzed by combined lipases: Optimization and kinetics. AIChE J. 2010, 56, 1659–1665. [Google Scholar] [CrossRef]
- Liu, Y.; Wu, H.; Yan, Y.; Dong, L.; Zhu, M.; Liang, P. Lipase-catalyzed transesterification for biodiesel production in ionic liquid [Emim]Tfo. Int. J. Green Energy 2013, 10, 63–71. [Google Scholar] [CrossRef]
- Tongboriboon, K.; Cheirsilp, B.; H-Kittikun, A. Mixed lipases for efficient enzymatic synthesis of biodiesel from used palm oil and ethanol in a solvent-free system. J. Mol. Catal. B Enzym. 2010, 67, 52–59. [Google Scholar] [CrossRef]
- Banerjee, A.; Singh, V.; Solanki, K.; Mukherjee, J.; Gupta, M. Combi-protein coated microcrystals of lipases for production of biodiesel from oil from spent coffee grounds. Sustain. Chem. Process. 2013, 1, 14. [Google Scholar] [CrossRef] [Green Version]
- Babaki, M.; Yousefi, M.; Habibi, Z.; Mohammadi, M. Process optimization for biodiesel production from waste cooking oil using multi-enzyme systems through response surface methodology. Renew. Energy 2017, 105, 465–472. [Google Scholar] [CrossRef] [Green Version]
- Poppe, J.K.; Matte, C.R.; de Freitas, V.O.; Fernandez-Lafuente, R.; Rodrigues, R.C.; Záchia Ayub, M.A. Enzymatic synthesis of ethyl esters from waste oil using mixtures of lipases in a plug-flow packed-bed continuous reactor. Biotechnol. Prog. 2018, 34, 952–959. [Google Scholar] [CrossRef]
- Araki, C.A.; Marcucci, S.M.P.; da Silva, L.S.; Maeda, C.H.; Arroyo, P.A.; Zanin, G.M. Effects of a combination of lipases immobilised on desilicated and thiol-modified ZSM-5 for the synthesis of ethyl esters from macauba pulp oil in a solvent-free system. Appl. Catal. A Gen. 2018, 562, 241–249. [Google Scholar] [CrossRef]
- Poppe, J.K.; Matte, C.R.; Fernandez-Lafuente, R.; Rodrigues, R.C.; Ayub, M.A.Z. Transesterification of waste frying oil and soybean oil by combi-lipases under ultrasound-assisted reactions. Appl. Biochem. Biotechnol. 2018, 186, 576–589. [Google Scholar] [CrossRef]
- de Freitas, V.O.; Matte, C.R.; Poppe, J.K.; Rodrigues, R.C.; Ayub, M.A.Z. Ultrasound-assisted transesterification of soybean oil using combi-lipase biocatalysts. Braz. J. Chem. Eng. 2019, 36, 995–1005. [Google Scholar] [CrossRef] [Green Version]
- Sánchez-Bayo, A.; Morales, V.; Rodríguez, R.; Vicente, G.; Bautista, L. Biodiesel production (FAEEs) by heterogeneous combi-lipase biocatalysts using wet extracted lipids from microalgae. Catalysts 2019, 9, 296. [Google Scholar] [CrossRef] [Green Version]
- Ramos, M.D.; Miranda, L.P.; Fernandez-Lafuente, R.; Kopp, W.; Tardioli, P.W. Improving the yields and reaction rate in the ethanolysis of soybean oil by using mixtures of lipase CLEAs. Molecules 2019, 24, 4392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schoevaart, R.; Wolbers, M.W.; Golubovic, M.; Ottens, M.; Kieboom, A.P.G.; van Rantwijk, F.; van der Wielen, L.A.M.; Sheldon, R.A. Preparation, optimization, and structures of cross-linked enzyme aggregates (CLEAs). Biotechnol. Bioeng. 2004, 87, 754–762. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Van Rantwijk, F.; Sheldon, R.A. Cross-linked enzyme aggregates: A simple and effective method for the immobilization of penicillin acylase. Org. Lett. 2000, 2, 1361–1364. [Google Scholar] [CrossRef]
- Chmura, A.; Rustler, S.; Paravidino, M.; Van Rantwijk, F.; Stolz, A.; Sheldon, R.A. The combi-CLEA approach: Enzymatic cascade synthesis of enantiomerically pure (S)-mandelic acid. Tetrahedron Asymmetry 2013, 24, 1225–1232. [Google Scholar] [CrossRef]
- Chung, C.-F.; Lin, S.-C.; Juang, T.-Y.; Liu, Y.-C. Shaking rate during production affects the activity of Escherichia coli surface-displayed Candida antarctica lipase A. Catalysts 2020, 10, 382. [Google Scholar] [CrossRef]
- Xu, L.; Xiao, X.; Wang, F.; He, Y.; Yang, X.; Hu, J.; Feng, Z.; Yan, Y. Surface-displayed thermostable Candida rugosa lipase 1 for docosahexaenoic acid enrichment. Appl. Biochem. Biotechnol. 2020, 190, 218–231. [Google Scholar] [CrossRef]
- Han, Z.; Han, S.; Zheng, S.; Lin, Y. Enhancing thermostability of a Rhizomucor miehei lipase by engineering a disulfide bond and displaying on the yeast cell surface. Appl. Microbiol. Biotechnol. 2009, 85, 117–126. [Google Scholar] [CrossRef]
- Matsumoto, T.; Fukuda, H.; Ueda, M.; Tanaka, A.; Kondo, A. Construction of yeast strains with high cell surface lipase activity by using novel display systems based on the Flo1p flocculation functional domain. Appl. Environ. Microbiol. 2002, 68, 4517–4522. [Google Scholar] [CrossRef] [Green Version]
- Jin, Z.; Han, S.-Y.; Zhang, L.; Zheng, S.-P.; Wang, Y.; Lin, Y. Combined utilization of lipase-displaying Pichia pastoris whole-cell biocatalysts to improve biodiesel production in co-solvent media. Bioresour. Technol. 2013, 130, 102–109. [Google Scholar] [CrossRef]
- Su, F.; Li, G.L.; Fan, Y.L.; Yan, Y.J. Enhancing biodiesel production via a synergic effect between immobilized Rhizopus oryzae lipase and Novozym 435. Fuel Process. Technol. 2015, 137, 298–304. [Google Scholar] [CrossRef]
- Jafarian Asl, P.; Niazmand, R.; Jahani, M. Theoretical and experimental assessment of supercritical CO2 in the extraction of phytosterols from rapeseed oil deodorizer distillates. J. Food Eng. 2020, 269, 109748. [Google Scholar] [CrossRef]
- Shao, P.; Jiang, S.T.; Ying, Y.J. Optimization of molecular distillation for recovery of tocopherol from rapeseed oil deodorizer distillate using response surface and artificial neural network models. Food Bioprod. Process. 2007, 85, 85–92. [Google Scholar] [CrossRef]
- Zhu, Z.N.; Xu, L.; Zhang, H.J.; Yan, Y.J. Efficient production of biodiesel from rapeseed oil deodorizer distillate: One-pot esterification and transesterfication with compound lipases. Appl. Mech. Mater. 2013, 291–294, 267–275. [Google Scholar] [CrossRef]
- Pedro, K.; Parreira, J.; Correia, I.; Henriques, C.; Langone, M. Enzymatic biodiesel synthesis from acid oil using a lipase mixture. Quim. Nova 2017, 41, 284–291. [Google Scholar] [CrossRef]
- Ibrahim, N.A.; Guo, Z.; Xu, X. Enzymatic interesterification of palm stearin and coconut oil by a dual lipase system. J. Am. Oil Chem. Soc. 2008, 85, 37–45. [Google Scholar] [CrossRef]
- Pande, G.; Sabir, J.S.M.; Baeshen, N.A.; Akoh, C.C. Enzymatic synthesis of extra virgin olive oil based infant formula fat analogues containing ARA and DHA: One-stage and two-stage syntheses. J. Agric. Food Chem. 2013, 61, 10590–10598. [Google Scholar] [CrossRef]
- Mat Radzi, S.; Abd Rahman, N.J.; Mohd Noor, H.; Basri, M. High yield synthesis of kojic ester using dual enzymes system and their antibacterial activity. Key Eng. Mater. 2013, 594–595, 362–369. [Google Scholar] [CrossRef]
- Mat Radzi, S.; Mohd Zulhilmi, M.N.; Mohd Noor, H.; Mohd Rehan, M. Preliminary study on the synthesis of kojic acid monooleate using dual enzymes system. AIP Conf. Proc. 2018, 2030, 020232. [Google Scholar]
- Cui, C.; Zhang, Z.; Zeng, Q.; Chen, B. Insight into the synthesis of isosorbide diester plasticizer using immobilized lipases. RSC Adv. 2016, 6, 108180–108186. [Google Scholar] [CrossRef]
- van Buijtenen, J.; van As, B.A.C.; Verbruggen, M.; Roumen, L.; Vekemans, J.A.J.M.; Pieterse, K.; Hilbers, P.A.J.; Hulshof, L.A.; Palmans, A.R.A.; Meijer, E.W. Switching from S- to R-selectivity in the Candida antarctica lipase B-catalyzed ring-opening of ω-methylated lactones: Tuning polymerizations by ring size. J. Am. Chem. Soc. 2007, 129, 7393–7398. [Google Scholar] [CrossRef]
- Fernandez-Lorente, G.; Palomo, J.M.; Cabrera, Z.; Fernandez-Lafuente, R.; Guisán, J.M. Improved catalytic properties of immobilized lipases by the presence of very low concentrations of detergents in the reaction medium. Biotechnol. Bioeng. 2007, 97, 242–250. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, T.; Kanamaru, R.; Watanabe, K.; Kamitanaka, T.; Harada, T.; Nakamura, K. Control of enantioselectivity of lipase-catalyzed esterification in supercritical carbon dioxide by tuning the pressure and temperature. Tetrahedron Asymmetry 2003, 14, 2087–2091. [Google Scholar] [CrossRef]
- Quirós, M.; Parker, M.C.; Turner, N.J. Tuning lipase enantioselectivity in organic media using solid-state buffers. J. Org. Chem. 2001, 66, 5074–5079. [Google Scholar] [CrossRef] [PubMed]
- Odaneth, A.A.; Vadgama, R.N.; Bhat, A.D.; Lali, A.M. Tuning lipase reaction for production of fatty acids from oil. Appl. Biochem. Biotechnol. 2016, 180, 504–515. [Google Scholar] [CrossRef]
- Acevedo-Rocha, C.G.; Reetz, M.T. Tuning lipase activity with perfluoro carboxylic acids as additives. Catal. Sci. Technol. 2012, 2, 1553–1555. [Google Scholar] [CrossRef]
- Palomo, J.M.; Fernandez-Lorente, G.; Mateo, C.; Ortiz, C.; Fernandez-Lafuente, R.; Guisan, J.M. Modulation of the enantioselectivity of lipases via controlled immobilization and medium engineering: Hydrolytic resolution of mandelic acid esters. Enzyme Microb. Technol. 2002, 31, 775–783. [Google Scholar] [CrossRef]
- Fernández-Lorente, G.; Palomo, J.M.; Cabrera, Z.; Guisán, J.M.; Fernández-Lafuente, R. Specificity enhancement towards hydrophobic substrates by immobilization of lipases by interfacial activation on hydrophobic supports. Enzyme Microb. Technol. 2007, 41, 565–569. [Google Scholar] [CrossRef]
- Palomo, J.M.; Muñoz, G.; Fernández-Lorente, G.; Mateo, C.; Fuentes, M.; Guisan, J.M.; Fernández-Lafuente, R. Modulation of Mucor miehei lipase properties via directed immobilization on different hetero-functional epoxy resins: Hydrolytic resolution of (R,S)-2-butyroyl-2-phenylacetic acid. J. Mol. Catal. B Enzym. 2003, 21, 201–210. [Google Scholar] [CrossRef]
- Wilson, L.; Palomo, J.M.; Fernández-Lorente, G.; Illanes, A.; Guisán, J.M.; Fernández-Lafuente, R. Improvement of the functional properties of a thermostable lipase from Alcaligenes sp. via strong adsorption on hydrophobic supports. Enzyme Microb. Technol. 2006, 38, 975–980. [Google Scholar] [CrossRef]
- Fernandez-Lorente, G.; Cabrera, Z.; Godoy, C.; Fernandez-Lafuente, R.; Palomo, J.M.; Guisan, J.M. Interfacially activated lipases against hydrophobic supports: Effect of the support nature on the biocatalytic properties. Process Biochem. 2008, 43, 1061–1067. [Google Scholar] [CrossRef]
- Cabrera, Z.; Fernandez-Lorente, G.; Fernandez-Lafuente, R.; Palomo, J.M.; Guisan, J.M. Novozym 435 displays very different selectivity compared to lipase from Candida antarctica B adsorbed on other hydrophobic supports. J. Mol. Catal. B Enzym. 2009, 57, 171–176. [Google Scholar] [CrossRef]
- Hernandez, K.; Garcia-Galan, C.; Fernandez-Lafuente, R. Simple and efficient immobilization of lipase B from Candida antarctica on porous styrene-divinylbenzene beads. Enzyme Microb. Technol. 2011, 49, 72–78. [Google Scholar] [CrossRef] [PubMed]
- Abreu Silveira, E.; Moreno-Perez, S.; Basso, A.; Serban, S.; Pestana Mamede, R.; Tardioli, P.W.; Sanchez Farinas, C.; Rocha-Martin, J.; Fernandez-Lorente, G.; Guisan, J.M. Modulation of the regioselectivity of Thermomyces lanuginosus lipase via biocatalyst engineering for the ethanolysis of oil in fully anhydrous medium. BMC Biotechnol. 2017, 17, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Abreu Silveira, E.; Moreno-Perez, S.; Basso, A.; Serban, S.; Pestana-Mamede, R.; Tardioli, P.W.; Farinas, C.S.; Castejon, N.; Fernandez-Lorente, G.; Rocha-Martin, J.; et al. Biocatalyst engineering of Thermomyces Lanuginosus lipase adsorbed on hydrophobic supports: Modulation of enzyme properties for ethanolysis of oil in solvent-free systems. J. Biotechnol. 2019, 289, 126–134. [Google Scholar] [CrossRef] [PubMed]
- Toro, E.C.; Rodríguez, D.F.; Morales, N.; García, L.M.; Godoy, C.A. Novel combi-lipase systems for fatty acid ethyl esters production. Catalysts 2019, 9, 546. [Google Scholar] [CrossRef] [Green Version]
- Tacias-Pascacio, V.G.; Virgen-Ortíz, J.J.; Jiménez-Pérez, M.; Yates, M.; Torrestiana-Sanchez, B.; Rosales-Quintero, A.; Fernandez-Lafuente, R. Evaluation of different lipase biocatalysts in the production of biodiesel from used cooking oil: Critical role of the immobilization support. Fuel 2017, 200, 1–10. [Google Scholar] [CrossRef]
- Tacias-Pascacio, V.G.; Torrestiana-Sánchez, B.; Dal Magro, L.; Virgen-Ortíz, J.J.; Suárez-Ruíz, F.J.; Rodrigues, R.C.; Fernandez-Lafuente, R. Comparison of acid, basic and enzymatic catalysis on the production of biodiesel after RSM optimization. Renew. Energy 2019, 135, 1–9. [Google Scholar] [CrossRef]
- Zhang, G.; Quin, M.B.; Schmidt-Dannert, C. Self-assembling protein scaffold system for easy in vitro coimmobilization of biocatalytic cascade enzymes. ACS Catal. 2018, 8, 5611–5620. [Google Scholar] [CrossRef]
- Talekar, S.; Pandharbale, A.; Ladole, M.; Nadar, S.; Mulla, M.; Japhalekar, K.; Pattankude, K.; Arage, D. Carrier free co-immobilization of alpha amylase, glucoamylase and pullulanase as combined cross-linked enzyme aggregates (combi-CLEAs): A tri-enzyme biocatalyst with one pot starch hydrolytic activity. Bioresour. Technol. 2013, 147, 269–275. [Google Scholar] [CrossRef]
- Li, Y.; Wen, L.; Tan, T.; Lv, Y. Sequential co-immobilization of enzymes in metal-organic frameworks for efficient biocatalytic conversion of adsorbed CO2 to formate. Front. Bioeng. Biotechnol. 2019, 7, 394. [Google Scholar] [CrossRef] [Green Version]
- Lopez-Gallego, F.; Schmidt-Dannert, C. Multi-enzymatic synthesis. Curr. Opin. Chem. Biol. 2010, 14, 174–183. [Google Scholar] [CrossRef] [PubMed]
- Velasco-Lozano, S.; Benítez-Mateos, A.I.; López-Gallego, F. Co-immobilized phosphorylated cofactors and enzymes as self-sufficient heterogeneous biocatalysts for chemical processes. Angew. Chemie Int. Ed. 2017, 56, 771–775. [Google Scholar] [CrossRef] [PubMed]
- Schmidt-Dannert, C.; Lopez-Gallego, F. A roadmap for biocatalysis – functional and spatial orchestration of enzyme cascades. Microb. Biotechnol. 2016, 9, 601–609. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmid-Dannert, C.; López-Gallego, F. Advances and opportunities for the design of self-sufficient and spatially organized cell-free biocatalytic systems. Curr. Opin. Chem. Biol. 2019, 49, 97–104. [Google Scholar] [CrossRef] [PubMed]
- López-Gallego, F.; Jackson, E.; Betancor, L. Heterogeneous systems biocatalysis: The path to the fabrication of self-sufficient artificial metabolic cells. Chem. A Eur. J. 2017, 23, 17841–17849. [Google Scholar] [CrossRef] [PubMed]
- Velasco-Lozano, S.; López-Gallego, F. Wiring step-wise reactions with immobilized multi-enzyme systems. Biocatal. Biotransform. 2018, 36, 184–194. [Google Scholar] [CrossRef]
- Peirce, S.; Virgen-Ortíz, J.J.; Tacias-Pascacio, V.G.; Rueda, N.; Bartolome-Cabrero, R.; Fernandez-Lopez, L.; Russo, M.E.; Marzocchella, A.; Fernandez-Lafuente, R. Development of simple protocols to solve the problems of enzyme coimmobilization. Application to coimmobilize a lipase and a β-galactosidase. RSC Adv. 2016, 6, 61707–61715. [Google Scholar] [CrossRef]
- Lee, J.H.; Kim, S.B.; Yoo, H.Y.; Lee, J.H.; Park, C.; Han, S.O.; Kim, S.W. Kinetic modeling of biodiesel production by mixed immobilized and co-immobilized lipase systems under two pressure conditions. Korean J. Chem. Eng. 2013, 30, 1272–1276. [Google Scholar] [CrossRef]
- Lee, J.H.; Kim, S.B.; Yoo, H.Y.; Lee, J.H.; Han, S.O.; Park, C.; Kim, S.W. Co-immobilization of Candida rugosa and Rhyzopus oryzae lipases and biodiesel production. Korean J. Chem. Eng. 2013, 30, 1335–1338. [Google Scholar] [CrossRef]
- Lee, J.H.; Lee, J.H.; Kim, D.S.; Yoo, H.Y.; Park, C.; Kim, S.W. Biodiesel production by lipases co-immobilized on the functionalized activated carbon. Bioresour. Technol. Rep. 2019, 7, 100248. [Google Scholar] [CrossRef]
- Shahedi, M.; Yousefi, M.; Habibi, Z.; Mohammadi, M.; As’habi, M.A. Co-immobilization of Rhizomucor miehei lipase and Candida antarctica lipase B and optimization of biocatalytic biodiesel production from palm oil using response surface methodology. Renew. Energy 2019, 141, 847–857. [Google Scholar] [CrossRef]
- Kreiner, M.; Parker, M.C. High-activity biocatalysts in organic media: Solid-state buffers as the immobilisation matrix for protein-coated microcrystals. Biotechnol. Bioeng. 2004, 87, 24–33. [Google Scholar] [CrossRef] [PubMed]
- Shah, S.; Sharma, A.; Gupta, M.N. Cross-linked protein-coated microcrystals as biocatalysts in non-aqueous solvents. Biocatal. Biotransform. 2008, 26, 266–271. [Google Scholar] [CrossRef]
- Yildirim, D.; Toprak, A.; Alagöz, D.; Tukel, S.S. Protein-coated microcrystals of Prunus armeniaca hydroxynitrile lyase: An effective and recyclable biocatalyst for synthesis of (R)-mandelonitrile. Chem. Pap. 2019, 73, 185–193. [Google Scholar] [CrossRef]
- Yan, Y.; Xu, L.; Dai, M. A synergetic whole-cell biocatalyst for biodiesel production. RSC Adv. 2012, 2, 6170. [Google Scholar] [CrossRef]
- Hepziba Suganthi, S.; Swathi, K.V.; Biswas, R.; Basker, S.; Ramani, K. Co-immobilization of multiple enzymes onto surface-functionalized magnetic nanoparticle for the simultaneous hydrolysis of multiple substrates containing industrial wastes. Appl. Nanosci. 2019, 9, 1439–1457. [Google Scholar] [CrossRef]
- Dalal, S.; Kapoor, M.; Gupta, M.N. Preparation and characterization of combi-CLEAs catalyzing multiple non-cascade reactions. J. Mol. Catal. B Enzym. 2007, 44, 128–132. [Google Scholar] [CrossRef]
- Zaak, H.; Kornecki, J.F.; Siar, E.-H.; Fernandez-Lopez, L.; Corberán, V.C.; Sassi, M.; Fernandez-Lafuente, R. Coimmobilization of enzymes in bilayers using PEI as a glue to reuse the most stable enzyme: Preventing pei release during inactivated enzyme desorption. Process Biochem. 2017, 61, 95–101. [Google Scholar] [CrossRef]
- Arana-Peña, S.; Mendez-Sanchez, C.; Rios, N.S.; Ortiz, C.; Gonçalves, L.R.B.; Fernandez-Lafuente, R. New applications of glyoxyl-octyl agarose in lipases co-immobilization: Strategies to reuse the most stable lipase. Int. J. Biol. Macromol. 2019, 131, 989–997. [Google Scholar] [CrossRef]
- Rios, N.S.; Arana-Peña, S.; Mendez-Sanchez, C.; Ortiz, C.; Gonçalves, L.R.B.; Fernandez-Lafuente, R. Reuse of lipase from Pseudomonas fluorescens via its step-by-step coimmobilization on glyoxyl-octyl agarose beads with least stable lipases. Catalysts 2019, 9, 487. [Google Scholar] [CrossRef] [Green Version]
- Rios, N.S.; Arana-Peña, S.; Mendez-Sanchez, C.; Lokha, Y.; Cortes-Corberan, V.; Gonçalves, L.R.B.; Fernandez-Lafuente, R. Increasing the enzyme loading capacity of porous supports by a layer-by-layer immobilization strategy using PEI as glue. Catalysts 2019, 9, 576. [Google Scholar] [CrossRef] [Green Version]
- Arana-Peña, S.; Rios, N.S.; Mendez-Sanchez, C.; Lokha, Y.; Gonçalves, L.R.B.; Fernández-Lafuente, R. Use of polyethylenimine to produce immobilized lipase multilayers biocatalysts with very high volumetric activity using octyl-agarose beads: Avoiding enzyme release during multilayer production. Enzyme Microb. Technol. 2020, 137, 109535. [Google Scholar] [CrossRef] [PubMed]
- Arana-Peña, S.; Rios, N.S.; Mendez-Sanchez, C.; Lokha, Y.; Carballares, D.; Gonçalves, L.R.B.; Fernandez-Lafuente, R. Coimmobilization of different lipases: Simple layer by layer enzyme spatial ordering. Int. J. Biol. Macromol. 2019, 145, 856–864. [Google Scholar] [CrossRef] [PubMed]
- Arana-Peña, S.; Carballares, D.; Fernandez-Lafuente, R. Multi-combilipases: Coimmobilizing lipases with very different stabilities combining immobilization via interfacial activation and ion exchange: The reuse of the most stable coimmobilized enzymes after inactivation of the least stable enzymes. Catalysts. In preparation.




























© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arana-Peña, S.; Carballares, D.; Berenguer-Murcia, Á.; Alcántara, A.R.; Rodrigues, R.C.; Fernandez-Lafuente, R. One Pot Use of Combilipases for Full Modification of Oils and Fats: Multifunctional and Heterogeneous Substrates. Catalysts 2020, 10, 605. https://0-doi-org.brum.beds.ac.uk/10.3390/catal10060605
Arana-Peña S, Carballares D, Berenguer-Murcia Á, Alcántara AR, Rodrigues RC, Fernandez-Lafuente R. One Pot Use of Combilipases for Full Modification of Oils and Fats: Multifunctional and Heterogeneous Substrates. Catalysts. 2020; 10(6):605. https://0-doi-org.brum.beds.ac.uk/10.3390/catal10060605
Chicago/Turabian StyleArana-Peña, Sara, Diego Carballares, Ángel Berenguer-Murcia, Andrés R. Alcántara, Rafael C. Rodrigues, and Roberto Fernandez-Lafuente. 2020. "One Pot Use of Combilipases for Full Modification of Oils and Fats: Multifunctional and Heterogeneous Substrates" Catalysts 10, no. 6: 605. https://0-doi-org.brum.beds.ac.uk/10.3390/catal10060605