The Length Polymorphism of the 9th Intron in the Avian CHD1 Gene Allows Sex Determination in Some Species of Palaeognathae
Abstract
:1. Introduction
2. Materials and Methods
2.1. Biological Samples and DNA Extraction
2.2. DNA Amplification
3. Results and Discussion
3.1. CHD1iA and NIPBLi16 Markers Are Not Suitable for Sex Determination of Palaeognathous Birds
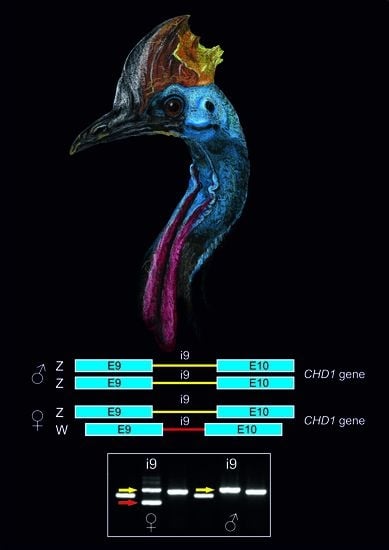
3.2. The CHD1i9 Marker Enables Sex Determination in Selected Species of Palaeognathous Birds
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dyke, G.J. The evolutionary radiation of modern birds: Systematics and patterns of diversification. Geol. J. 2001, 36, 305–315. [Google Scholar] [CrossRef]
- McInerney, P.L.; Lee, M.S.Y.; Clement, A.M.; Worthy, T.H. The phylogenetic significance of the morphology of the syrinx, hyoid and larynx, of the southern cassowary, Casuarius casuarius (Aves, Palaeognathae). BMC Evol. Biol. 2019, 19, 233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dyke, G.J.; Leonard, L.M. Palaeognathae. In eLS; John Wiley & Sons, Ltd.: Chichester, UK, 2012. [Google Scholar] [CrossRef]
- Van Tuinen, M.; Sibley, C.G.; Hedges, S.B. The early history of modern birds inferred from DNA sequences of nuclear and mitochondrial ribosomal genes. Mol. Biol. Evol. 2000, 17, 451–457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Widrig, K.; Field, D.J. The evolution and fossil record of Palaeognathous birds (Neornithes: Palaeognathae). Diversity 2022, 14, 105. [Google Scholar] [CrossRef]
- Gill, F.; Donsker, D.; Rasmussen, P. (Eds.) IOC World Bird List (v 11.1); IOC: Lausanne, Switzerland, 2021. [Google Scholar] [CrossRef]
- Claramunt, S.; Cracraft, J. A new time tree reveals Earth history’s imprint on the evolution of modern birds. Sci. Adv. 2015, 1, e1501005. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prum, R.O.; Berv, J.S.; Dornburg, A.; Field, D.J.; Townsend, J.P.; Lemmon, E.M.; Lemmon, A.R. A comprehensive phylogeny of birds (Aves) using targeted next-generation DNA sequencing. Nature 2015, 526, 569–573. [Google Scholar] [CrossRef] [PubMed]
- Yonezawa, T.; Segawa, T.; Mori, H.; Campos, P.; Hongoh, Y.; Endo, H.; Akiyoshi, A.; Kohno, N.; Nishida, S.; Wu, J.; et al. Phylogenomics and morphology of extinct paleognaths reveal the origin and evolution of the ratites. Curr. Biol. 2016, 27, 68–77. [Google Scholar] [CrossRef] [PubMed]
- Dawson, D.A.; dos Remedios, N.; Horsburgh, G.J. A new marker based on the avian spindlin gene that is able to sex most birds, including species problematic to sex with CHD markers. Zoo Biol. 2016, 35, 533–545. [Google Scholar] [CrossRef] [PubMed]
- Richner, H. Avian laparoscopy as a field technique for sexing birds and an assessment of its effect on wild birds. J. Field Ornithol. 1989, 60, 137–142. [Google Scholar]
- Redelman, D.; Fleury, S.A.; Garner, D.L. Flow cytometry for sexing birds. Trends Ecol. Evol. 1997, 12, 489. [Google Scholar] [CrossRef]
- Griffiths, R. Sex identification in birds. Semin. Avian Exot. Pet Med. 2000, 9, 14–26. [Google Scholar] [CrossRef]
- Kroczak, A.; Wołoszyńska, M.; Wierzbicki, H.; Kurkowski, M.; Grabowski, K.A.; Piasecki, T.; Galosi, L.; Urantówka, A.D. New bird sexing strategy developed in the order psittaciformes involves multiple markers to avoid sex misidentification: De-bunked myth of the universal DNA marker. Genes 2021, 12, 878. [Google Scholar] [CrossRef] [PubMed]
- Yuda, P.; Saputra, A.W. Eggshell membrane for DNA sexing of the endangered Maleo (Macrocephalon maleo). F1000Research 2021, 9, 599. [Google Scholar] [CrossRef] [PubMed]
- Ushine, N.; Kato, T.; Hayama, S.-I. Sex identification in Japanese birds using droppings as a source of DNA. Bull. Jpn. Bird Band. Assoc. 2016, 28, 51–70. [Google Scholar] [CrossRef] [Green Version]
- Fridolfsson, A.-K.; Cheng, H.; Copeland, N.G.; Jenkins, N.A.; Liu, H.-C.; Raudsepp, T.; Woodage, T.; Chowdhary, B.; Halverson, J.; Ellegren, H. Evolution of the avian sex chromosomes from an ancestral pair of autosomes. Proc. Natl. Acad. Sci. USA 1998, 95, 8147–8152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bellott, D.W.; Skaletsky, H.; Pyntikova, T.; Mardis, E.R.; Graves, T.; Kremitzki, C.; Brown, L.G.; Rozen, S.G.; Warren, W.C.; Wilson, R.K.; et al. Convergent evolution of chicken Z and human X chromosomes by expansion and gene acquisition. Nature 2010, 466, 612–616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shetty, S.; Griffin, D.K.; Graves, J.A.M. Comparative painting reveals strong chromosome homology over 80 million years of bird evolution. Chromosom. Res. 1999, 7, 289–295. [Google Scholar] [CrossRef] [PubMed]
- Okuno, M.; Mizushima, S.; Kuroiwa, A.; Itoh, T. Analysis of sex chromosome evolution in the clade Palaeognathae from phased genome assembly. Genome Biol. Evol. 2021, 13, evab242. [Google Scholar] [CrossRef] [PubMed]
- Tsuda, Y.; Nishida-Umehara, C.; Ishijima, J.; Yamada, K.; Matsuda, Y. Comparison of the Z and W sex chromosomal archi-tectures in elegant crested tinamou (Eudromia elegans) and ostrich (Struthio camelus) and the process of sex chromosome differentiation in palaeognathous birds. Chromosoma 2007, 116, 159–173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mank, J.E.; Ellegren, H. Parallel divergence and degradation of the avian W sex chromosome. Trends Ecol. Evol. 2007, 22, 389–391. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Zhang, J.; Bachtrog, D.; An, N.; Huang, Q.; Jarvis, E.D.; Gilbert, M.T.P.; Zhang, G. Complex evolutionary trajectories of sex chromosomes across bird taxa. Science 2014, 346, 6215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- International Chicken Genome Sequencing Consortium. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 2004, 432, 695–716. [Google Scholar] [CrossRef] [PubMed]
- Lessells, C.M.; Mateman, A.C. Sexing birds using random amplified polymorphic DNA (RAPD) markers. Mol. Ecol. 1998, 7, 187–195. [Google Scholar] [CrossRef]
- Smeds, L.; Warmuth, V.; Bolivar, P.; Uebbing, S.; Burri, R.; Suh, A.; Nater, A.; Bureš, S.; Garamszegi, L.Z.; Hogner, S.; et al. Evolutionary analysis of the female-specific avian W chromosome. Nat. Commun. 2015, 6, 7330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morinha, F.; Cabral, J.; Bastos, E. Molecular sexing of birds: A comparative review of polymerase chain reaction (PCR)-based methods. Theriogenology 2012, 78, 703–714. [Google Scholar] [CrossRef] [PubMed]
- Griffiths, R.; Daan, S.; Dijkstra, C. Sex identification in birds using two CHD genes. Proc. R. Soc. B Boil. Sci. 1996, 263, 1251–1256. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.-C.; Chen, C.-T.; Lee, H.-Y.; Li, S.-H.; Lir, J.-T.; Chin, S.-C.; Pu, C.-E.; Wang, C.-H. Sexing a wider range of avian species based on twoCHD1 introns with a unified reaction condition. Zoo Biol. 2007, 26, 425–431. [Google Scholar] [CrossRef] [PubMed]
- Ong, A.H.; Vellayan, S. An evaluation of CHD-Specific primer sets for sex typing of birds from feathers. Zoo Biol. 2008, 27, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Tsai, L.-C.; Hwa, P.-Y.; Chan, C.-L.; Huang, A.; Chin, S.-C.; Wang, L.-C.; Lin, J.-T.; Linacre, A.; Hsieh, H.-M. A novel strategy for avian species and gender identification using the CHD gene. Mol. Cell. Probes 2010, 24, 27–31. [Google Scholar] [CrossRef] [PubMed]
- Suh, A.; Kriegs, J.O.; Brosius, J.; Schmitz, J. Retroposon insertions and the chronology of avian sex chromosome evolution. Mol. Biol. Evol. 2011, 28, 2993–2997. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Kloet, R.; De Kloet, S. Evolution of the spindlin gene in birds: Independent cessation of the recombination of sex chromosomes at the spindlin locus in neognathous birds and tinamous, a palaeognathous avian family. Genetica 2003, 119, 333–342. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Xue, F.; Li, L.; Li, X.; Yue, B.; Li, J. A triple-primer PCR approach for the sex identification of endangered Phasianidae birds. Eur. J. Wildl. Res. 2012, 58, 289–294. [Google Scholar] [CrossRef]
- Huynen, L.; Miles, J.; Lambert, D. Unusual electrophoretic mobility of a DNA fragment of the universal ‘non-ratite’ sexing marker CHD allows sexing of New Zealand’s endangered kiwi ratite Apteryx spp. Ibis 2006, 148, 167–168. [Google Scholar] [CrossRef]
- Kahn, N.W.; John, J.S.T.; Quinn, T.W. Chromosome-specific intron size differences in the avian CHD gene provide an efficient method for sex identification in birds. Auk 1998, 115, 1074–1078. [Google Scholar] [CrossRef] [Green Version]
- Vucicevic, M.; Stevanov-Pavlovic, M.; Stevanovic, J.; Bosnjak, J.; Gajic, B.; Aleksic, N.; Stanimirovic, Z. Sex determination in 58 bird species and evaluation of CHD gene as a universal molecular marker in bird sexing. Zoo Biol. 2013, 32, 269–276. [Google Scholar] [CrossRef] [PubMed]
- Han, J.-I.; Kim, J.-H.; Kim, S.; Park, S.-R.; Na, K.-J. A simple and improved DNA test for avian sex determination. Auk 2009, 126, 779–783. [Google Scholar] [CrossRef]
- Griffiths, R.; Double, M.C.; Orr, K.; Dawson, R.J.G. A DNA test to sex most birds. Mol. Ecol. 1998, 7, 1071–1075. [Google Scholar] [CrossRef] [PubMed]
- Fridolfsson, A.-K.; Ellegren, H. A simple and universal method for molecular sexing of non-ratite birds. J. Avian Biol. 1999, 30, 116–121. [Google Scholar] [CrossRef]
- Morinha, F.; Cabral, J.; Martins, S.; Cruz, E.; Alvura, N.; Nunes, P.; Direitinho, J.; Magalhães, P.C.L.F.D.S.; Bastos, E. (R)evolution in the molecular sexing of ratite birds: Identification and analysis of new candidate sex-linked markers. Avian Biol. Res. 2015, 8, 145–159. [Google Scholar] [CrossRef]
- Huynen, L.; Millar, C.D.; Lambert, D.M. A DNA test to sex ratite birds. Mol. Ecol. 2002, 11, 851–856. [Google Scholar] [CrossRef] [PubMed]
- Huynen, L.; Lambert, D.M.; McLennan, J.A.; Rickard, C.; Robertson, H.A. A DNA test for sex assignment in Kiwi (Apteryx spp.). Notornis 2003, 50, 231–233. [Google Scholar]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Koshiishi, Y.; Wada, K. A simplified protocol for molecular sexing in the emu (Dromaius novaehollandiae). Poult. Sci. 2018, 97, 1117–1119. [Google Scholar] [CrossRef] [PubMed]
- Kloet, S.R.D. Development of a CAPS (cleaved amplified polymorphics sequence) assay for sex identification of the emu (Dromaius novaehollandiae). Mol. Ecol. Notes 2001, 1, 273–275. [Google Scholar] [CrossRef]
- Dash, S.K.; Kumarasamy, P.; Malik, H.N.; Thiagarajan, V. Phylogenetic analysis of ratite bird emu (Dromaius novaehollandiae) using sex specific sequences. Indian J. Anim. Res. 2013, 47, 247–250. [Google Scholar]
- Maheshkumar, G.; Saravanan, R.; Mani, K.; Murali, N. Molecular marker based sex identification in ratite and non-ratite type of birds: A comparative study. Indian Vet. J. 2017, 94, 11–13. [Google Scholar]
- Fraire, H.R.; Martella, M.B. DNA test to sex the lesser rhea (Rhea pennata pennata). Br. Poult. Sci. 2006, 47, 375–377. [Google Scholar] [CrossRef] [PubMed]
- Alipanah, M.; Torkamanzehi, A.; Taghavi, H. Sex determination in ostrich (Struthio camelus) using DNA markers. Can. J. Anim. Sci. 2010, 90, 357–360. [Google Scholar] [CrossRef]
- Bello, N.; Sánchez, A. The identification of a sex-specific DNA marker in the ostrich using a random amplified polymorphic DNA (RAPD) assay. Mol. Ecol. 1999, 8, 667–669. [Google Scholar] [CrossRef]
- Medaglia, A.; Henrique-Silva, F. New PCR multiplexes for sex typing of ostriches. Braz. J. Biol. 2005, 65, 743–745. [Google Scholar] [CrossRef] [Green Version]
- Mine, O.; Mochakana, M.; Mpapho, T.; Motlhanka, D.; Kgwatalala, P. Application of a sex identification technique in juvenile ostriches and its potential application in Botswana. S. Afr. J. Anim. Sci. 2002, 32, 160–163. [Google Scholar] [CrossRef] [Green Version]
- Wu, C.P.; Horng, Y.M.; Yanga, K.T.; Huanga, C.W.; Huanga, M.C. Female-specific DNA sequences in ostriches. Mol. Cell. Probes. 2006, 20, 307–310. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.-W.; Huang, M.-C. A Simple and Rapid PCR-Based Method for Ostrich Sexing Using Micro Amounts of DNA. Reprod. Domest. Anim. 2008, 43, 643–646. [Google Scholar] [CrossRef] [PubMed]
- Hinckley, J.D.; Park, R.L.; Xiong, S.; Andersen, W.R.; Kooyman, D.L. Identification and Development of Sex Specific DNA Markers in the Ostrich Using Polymerase Chain Reaction. Int. J. Poult. Sci. 2005, 4, 663–669. [Google Scholar]
- Malagó, W.; Franco, H.M.; Matheucci, E.; Medaglia, A.; Henrique-Silva, F. Large scale sex typing of ostriches using DNA extracted from feathers. BMC Biotechnol. 2002, 2, 19. [Google Scholar] [CrossRef] [PubMed]
- Kloet, S.R.D. Molecular sex identification of tinamous with PCR using primers derived from the spindlin gene. Mol. Ecol. Notes 2002, 2, 465–466. [Google Scholar] [CrossRef]
- Ogata, M.; Koizumi, J.; Morikaku, K.; Matsumoto, R. Sexing, the Elegant Crested-Tinamou, Eudromia elegans, Using a Molecular Biological Method. Jpn. J. Zoo Wildl. Med. 2006, 11, 79–81. [Google Scholar] [CrossRef] [Green Version]
- Mazzoleni, S.; Němec, P.; Albrecht, T.; Lymberakis, P.; Kratochvíl, L.; Rovatsos, M. Long-term stability of sex chromosome gene content allows accurate qPCR-based molecular sexing across birds. Mol. Ecol. Resour. 2021, 21, 2013–2021. [Google Scholar] [CrossRef] [PubMed]


| Order | Species | Primers | Amplicons M/F | Impracticality | Utility | References |
|---|---|---|---|---|---|---|
| Apterygiformes | Apteryx mantelli | P2/P8 | 1/2 | separation in 6% polyacrylamide | + | [35] |
| Casuariiformes | Dromaius novaehollandiae | 1237L/1272H | 1/1 | any female specific band | − | [29,36] |
| Casuariiformes | Dromaius novaehollandiae | 2550F/2718R | 1/1 | any female specific band | − | [29,37] |
| Casuariiformes | Casuarius casuarius | 2550F/2718R | 1/1 | any female specific band | − | [37] |
| Rheiformes | Rhea americana | 2550F/2718R | 1/1 | any female specific band | − | [37] |
| Struthioniformes | Struthio camelus | P2/P8 + P0 | 1/2 | 3 primers are used | + + | [38] |
| Struthioniformes | Struthio camelus | 1237L/1272H | 1/1 | any female specific band | − | [29,36] |
| Struthioniformes | Struthio camelus | 2550F/2718R | 1/1 | any female specific band | − | [29,30] |
| Struthioniformes | Struthio camelus | P2/P8 | 1/1 | any female specific band | − | [39] |
| Species | Sex | Biosample | CHD1i9 | |
|---|---|---|---|---|
| Accession | Length (bp) | |||
| Apteryx rowi | Male | SAMN08476454 | NW_020450197.1 | 632 |
| Apteryx haastii | Male | SAMN08476452 | PTFD01000391.1 | 627 |
| Apteryx owenii | Male | SAMN08476453 | PTFC01000295.1 | 632 |
| Crypturellus cinnamomeus | Male | SAMN08476456 | PTEZ01000258.1 | 601 |
| Crypturellus soui | Female | SAMN12253745 | VWPX01020692.1 | 598 * |
| Crypturellus soui | Female | SAMN12253746 | VWPX01026815.1 | 220 ** |
| Crypturellus undulatus | Female | SAMN12253747 | VWPW01009891.1 | 601 * |
| Crypturellus undulatus | Female | SAMN12253746 | VWPW01026715.1 | 609 ** |
| Eudromia elegans | Male | SAMN08476458 | PTEX01000066.1 | 579 |
| Nothocercus nigrocapillus | Female | SAMN12253973 | WBNA01000102.1 | 590 |
| Nothoprocta pentlandii | Female | SAMN12253975 | VZSG01000618.1 | 596 |
| Nothoprocta perdicaria | Male | SAMN08476459 | NW_020455588.1 | 598 |
| Tinamus guttatus | Female | SAMN02316659 | NW_010585320.1 | 603 * |
| Tinamus guttatus | Female | SAMN02316660 | NW_010577858.1 | 583 ** |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kroczak, A.; Wierzbicki, H.; Urantówka, A.D. The Length Polymorphism of the 9th Intron in the Avian CHD1 Gene Allows Sex Determination in Some Species of Palaeognathae. Genes 2022, 13, 507. https://0-doi-org.brum.beds.ac.uk/10.3390/genes13030507
Kroczak A, Wierzbicki H, Urantówka AD. The Length Polymorphism of the 9th Intron in the Avian CHD1 Gene Allows Sex Determination in Some Species of Palaeognathae. Genes. 2022; 13(3):507. https://0-doi-org.brum.beds.ac.uk/10.3390/genes13030507
Chicago/Turabian StyleKroczak, Aleksandra, Heliodor Wierzbicki, and Adam Dawid Urantówka. 2022. "The Length Polymorphism of the 9th Intron in the Avian CHD1 Gene Allows Sex Determination in Some Species of Palaeognathae" Genes 13, no. 3: 507. https://0-doi-org.brum.beds.ac.uk/10.3390/genes13030507