Marine Bacterial Secondary Metabolites: A Treasure House for Structurally Unique and Effective Antimicrobial Compounds
Abstract
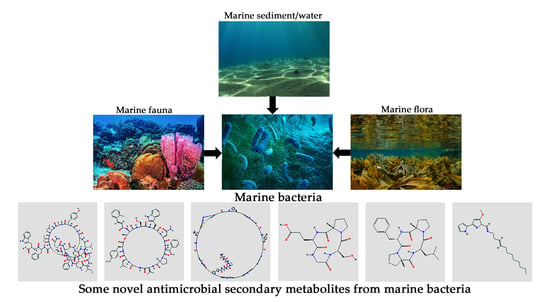
:1. Introduction
2. The Marine Environment: A Goldmine for Natural Product Research
3. Secondary Metabolites from Marine Bacteria: A Treasure House of Natural Bioactives
4. Antimicrobial Production by Different Phyla of Marine Bacteria
4.1. Antimicrobial Potential of Marine Actinobacteria
4.2. Antimicrobial Potential of Marine Bacteroidetes
4.3. Antimicrobial Potential of Marine Cyanobacteria
4.4. Antimicrobial Potential of Marine Firmicutes
4.5. Antimicrobial Potential of Marine Planctomycetes
4.6. Antimicrobial Potential of Marine Proteobacteria
5. Antimicrobial Potential of Marine Sediment-Derived Bacteria
6. Antimicrobial Potential of Marine Water-Derived Bacteria
7. Marine Fauna
7.1. Antimicrobial Potential of Bacteria Associated with Marine Sponges
7.2. Antimicrobial Potential of Bacteria Associated with Marine Corals
7.3. Antimicrobial Potential of Bacteria Associated with Marine Mollusks
8. Marine Flora
8.1. Antimicrobial Potential of Bacteria Associated with Marine Seaweeds
8.2. Antimicrobial Potential of Bacteria Associated with Marine Seagrasses
8.3. Antimicrobial Potential of Marine Bacteria Associated with Mangroves
9. Molecular Approaches for the Identification and Development of Novel Antimicrobial Agents from Marine Bacteria
10. Future Research Directions
11. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Doron, S.; Gorbach, S. Bacterial infections: Overview. Int. Encycl. Public Health 2008, 273–282. [Google Scholar] [CrossRef]
- Packiavathy, I.A.S.V.; Kannappan, A.; Thiyagarajan, S.; Srinivasan, R.; Jeyapragash, D.; Paul, J.B.J.; Velmurugan, P.; Ravi, A.V. AHL-Lactonase producing Psychrobacter sp. from Palk Bay sediment mitigates quorum sensing-mediated virulence production in Gram negative bacterial pathogens. Front. Microbiol. 2021, 12, 748. [Google Scholar] [CrossRef] [PubMed]
- Cole, L.; Kramer, P.R. Bacteria, virus, fungi, and infectious diseases. Hum. Physiol. Biochem. Basic Med. 2016, 193–196. [Google Scholar] [CrossRef]
- Boucher, H.W.; Talbot, G.H.; Bradley, J.S.; Edwards, J.E.; Gilbert, D.; Rice, L.B.; Scheld, M.; Spellberg, B.; Bartlett, J. Bad bugs, no drugs: No ESKAPE! An update from the Infectious Diseases Society of America. Clin. Infect. Dis. 2009, 48, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Mulani, M.S.; Kamble, E.E.; Kumkar, S.N.; Tawre, M.S.; Pardesi, K.R. Emerging strategies to combat ESKAPE pathogens in the era of antimicrobial resistance: A review. Front. Microbiol. 2019, 10, 539. [Google Scholar] [CrossRef]
- Santajit, S.; Indrawattana, N. Mechanisms of antimicrobial resistance in ESKAPE pathogens. BioMed Res. Int. 2016, 2016, 2475067. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schultz, F.; Anywar, G.; Tang, H.; Chassagne, F.; Lyles, J.T.; Garbe, L.-A.; Quave, C.L. Targeting ESKAPE pathogens with anti-infective medicinal plants from the Greater Mpigi region in Uganda. Sci. Rep. 2020, 10, 11935. [Google Scholar] [CrossRef]
- Brown, G.D.; Denning, D.W.; Gow, N.A.; Levitz, S.M.; Netea, M.G.; White, T.C. Hidden killers: Human fungal infections. Sci. Transl. Med. 2012, 4, 165rv13. [Google Scholar] [CrossRef] [Green Version]
- El-Hossary, E.M.; Cheng, C.; Hamed, M.M.; Hamed, A.N.E.-S.; Ohlsen, K.; Hentschel, U.; Abdelmohsen, U.R. Antifungal potential of marine natural products. Eur. J. Med. Chem. 2017, 126, 631–651. [Google Scholar] [CrossRef] [PubMed]
- Kullberg, B.J.; Arendrup, M.C. Invasive candidiasis. N. Engl. J. Med. 2015, 373, 1445–1456. [Google Scholar] [CrossRef] [Green Version]
- Park, S.J.; Mehrad, B. Innate immunity to Aspergillus species. Clin. Microbiol. Rev. 2009, 22, 535–551. [Google Scholar] [CrossRef] [Green Version]
- Dagenais, T.R.; Keller, N.P. Pathogenesis of Aspergillus fumigatus in invasive aspergillosis. Clin. Microbiol. Rev. 2009, 22, 447–465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.; Xiao, X.; Feng, H.; Hong, Z.; Li, M.; Tu, N.; Li, X.; Wang, K.; Bu, L. Ongoing COVID-19 Pandemic: A concise but updated comprehensive review. Curr. Microbiol. 2021, 78, 1718–1729. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.; Mishra, V. Environmental impacts of coronavirus disease 2019 (COVID-19). Bioresour. Technol. Rep. 2021, 15, 100744. [Google Scholar] [CrossRef]
- Vega, L.E.; Espinoza, L.R. Human immunodeficiency virus infection (HIV)–associated rheumatic manifestations in thepre-and post-HAART eras. Clin. Rheumatol. 2020, 39, 2515–2522. [Google Scholar] [CrossRef] [Green Version]
- Walsh, C. Antibiotics: Actions, Origins, Resistance; American Society for Microbiology (ASM): Washington, DC, USA, 2003. [Google Scholar]
- Srinivasan, R.; Santhakumari, S.; Poonguzhali, P.; Geetha, M.; Dyavaiah, M.; Lin, X. Bacterial biofilm inhibition: A focused review on recent therapeutic strategies for combating the biofilm mediated infections. Front. Microbiol. 2021, 12, 1106. [Google Scholar] [CrossRef]
- Li, Z.; Zhang, L.; Song, Q.; Wang, G.; Yang, W.; Tang, H.; Srinivasan, R.; Lin, L.; Lin, X. Proteomics analysis reveals bacterial antibiotics resistance mechanism mediated by ahslyA against Enoxacin in Aeromonas hydrophila. Front. Microbiol. 2021, 12, 1498. [Google Scholar]
- Kannappan, A.; Srinivasan, R.; Nivetha, A.; Annapoorani, A.; Pandian, S.K.; Ravi, A.V. Anti-virulence potential of 2-hydroxy-4-methoxybenzaldehyde against methicillin-resistant Staphylococcus aureus and its clinical isolates. Appl. Microbiol. Biotechnol. 2019, 103, 6747–6758. [Google Scholar] [CrossRef] [PubMed]
- Kannappan, A.; Sivaranjani, M.; Srinivasan, R.; Rathna, J.; Pandian, S.K.; Ravi, A.V. Inhibitory efficacy of geraniol on biofilm formation and development of adaptive resistance in Staphylococcus epidermidis RP62A. J. Med. Microbiol. 2017, 66, 1506–1515. [Google Scholar] [CrossRef]
- Li, Z.; Wang, Y.; Li, X.; Lin, Z.; Lin, Y.; Srinivasan, R.; Lin, X. The characteristics of antibiotic resistance and phenotypes in 29 outer-membrane protein mutant strains in Aeromonas hydrophila. Environ. Microbiol. 2019, 21, 4614–4628. [Google Scholar] [CrossRef]
- Podolsky, S.H. The evolving response to antibiotic resistance (1945–2018). Palgrave Commun. 2018, 4, 124. [Google Scholar] [CrossRef]
- Alexpandi, R.; Prasanth, M.I.; Ravi, A.V.; Balamurugan, K.; Durgadevi, R.; Srinivasan, R.; De Mesquita, J.F.; Pandian, S.K. Protective effect of neglected plant Diplocyclos palmatus on quorum sensing mediated infection of Serratia marcescens and UV-A induced photoaging in model Caenorhabditis elegans. J. Photochem. Photobiol. B Biol. 2019, 201, 111637. [Google Scholar] [CrossRef]
- World Health Organization. Global Antimicrobial Resistance Surveillance System: Manual for Early Implementation; World Health Organization: Geneva, Switzerland, 2015. [Google Scholar]
- De Oliveira, D.M.; Forde, B.M.; Kidd, T.J.; Harris, P.N.; Schembri, M.A.; Beatson, S.A.; Paterson, D.L.; Walker, M.J. Antimicrobial resistance in ESKAPE pathogens. Clin. Microbiol. Rev. 2020, 33, e00181-19. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, J. Tackling Drug-Resistant Infections Gobally: Final Report and Recommendations—The Review on Antimicrobial Resistance Chaired by Jim O’Neill; Wellcome Trust and HM Government: London, UK, 2016. [Google Scholar]
- Fu, Y.; Zhang, L.; Wang, G.; Lin, Y.; Ramanathan, S.; Yang, G.; Lin, W.; Lin, X. The LysR-type transcriptional regulator YeeY plays important roles in the regulatory of furazolidone resistance in Aeromonas hydrophila. Front. Microbiol. 2020, 11, 2208. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Ramanathan, S.; Chen, L.; Zeng, F.; Li, X.; Zhao, Y.; Lin, L.; Monaghan, S.J.; Lin, X.; Pang, H. Comparative transcriptomic analysis reveals the molecular mechanisms related to oxytetracycline-resistance in strains of Aeromonas hydrophila. Aquac. Rep. 2021, 21, 100812. [Google Scholar] [CrossRef]
- Li, W.; Zhao, Y.; Yu, J.; Lin, L.; Ramanathan, S.; Wang, G.; Lin, X.; Pang, H. TonB-dependent receptors affect the spontaneous oxytetracycline resistance evolution in Aeromonas hydrophila. J. Proteome Res. 2020, 20, 154–163. [Google Scholar] [CrossRef]
- Payne, D.J.; Michael, N.; Gwynn, D.J.H.; Pompliano, D.L.; Silver, L.L.; Lomovskaya, O.; Zgurskaya, H.I.; Totrov, M.; Watkins, W.J. The end of an era? Nature 2007, 6, 28. [Google Scholar]
- Koehn, F.E.; Carter, G.T. The evolving role of natural products in drug discovery. Nat. Rev. Drug Discov. 2005, 4, 206–220. [Google Scholar] [CrossRef]
- Liu, M.; El-Hossary, E.M.; Oelschlaeger, T.A.; Donia, M.S.; Quinn, R.J.; Abdelmohsen, U.R. Potential of marine natural products against drug-resistant bacterial infections. Lancet Infect. Dis. 2019, 19, e237–e245. [Google Scholar] [CrossRef]
- Choudhary, A.; Naughton, L.M.; Montánchez, I.; Dobson, A.D.; Rai, D.K. Current status and future prospects of marine natural products (MNPs) as antimicrobials. Mar. Drugs 2017, 15, 272. [Google Scholar] [CrossRef]
- Burgess, J.G. New and emerging analytical techniques for marine biotechnology. Curr. Opin. Biotechnol. 2012, 23, 29–33. [Google Scholar] [CrossRef] [PubMed]
- Martins, A.; Vieira, H.; Gaspar, H.; Santos, S. Marketed marine natural products in the pharmaceutical and cosmeceutical industries: Tips for success. Mar. Drugs 2014, 12, 1066–1101. [Google Scholar] [CrossRef] [Green Version]
- Blunt, J.W.; Copp, B.R.; Munro, M.H.; Northcote, P.T.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2011, 28, 196–268. [Google Scholar] [CrossRef]
- Encarnação, T.; Pais, A.A.; Campos, M.G.; Burrows, H.D. Cyanobacteria and microalgae: A renewable source of bioactive compounds and other chemicals. Sci. Prog. 2015, 98, 145–168. [Google Scholar] [CrossRef]
- Leal, M.C.; Munro, M.H.; Blunt, J.W.; Puga, J.; Jesus, B.; Calado, R.; Rosa, R.; Madeira, C. Biogeography and biodiscovery hotspots of macroalgal marine natural products. Nat. Prod. Rep. 2013, 30, 1380–1390. [Google Scholar] [CrossRef]
- Hegazy, M.E.F.; Mohamed, T.A.; Alhammady, M.A.; Shaheen, A.M.; Reda, E.H.; Elshamy, A.I.; Aziz, M.; Paré, P.W. Molecular architecture and biomedical leads of terpenes from red sea marine invertebrates. Mar. Drugs 2015, 13, 3154–3181. [Google Scholar] [CrossRef]
- Singh, R.P.; Kumari, P.; Reddy, C. Antimicrobial compounds from seaweeds-associated bacteria and fungi. Appl. Microbiol. Biotechnol. 2015, 99, 1571–1586. [Google Scholar] [CrossRef]
- Hou, X.-M.; Xu, R.-F.; Gu, Y.-C.; Wang, C.-Y.; Shao, C.-L. Biological and chemical diversity of coral-derived microorganisms. Curr. Med. Chem. 2015, 22, 3707–3762. [Google Scholar] [CrossRef]
- Srinivasan, R.; Santhakumari, S.; Ravi, A.V. In vitro antibiofilm efficacy of Piper betle against quorum sensing mediated biofilm formation of luminescent Vibrio harveyi. Microb. Pathog. 2017, 110, 232–239. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, R.; Devi, K.R.; Kannappan, A.; Pandian, S.K.; Ravi, A.V. Piper betle and its bioactive metabolite phytol mitigates quorum sensing mediated virulence factors and biofilm of nosocomial pathogen Serratia marcescens in vitro. J. Ethnopharmacol. 2016, 193, 592–603. [Google Scholar] [CrossRef] [PubMed]
- Kannappan, A.; Santhakumari, S.; Srinivasan, R.; Pandian, S.K.; Ravi, A.V. Hemidesmus indicus, a traditional medicinal plant, targets the adherence of multidrug-resistant pathogens to form biofilms. Biocatal. Agric. Biotechnol. 2019, 21, 101338. [Google Scholar] [CrossRef]
- Kannappan, A.; Gowrishankar, S.; Srinivasan, R.; Pandian, S.K.; Ravi, A.V. Antibiofilm activity of Vetiveria zizanioides root extract against methicillin-resistant Staphylococcus aureus. Microb. Pathog. 2017, 110, 313–324. [Google Scholar] [CrossRef]
- Ma, Y. Biotechnological potential of plant-microbe interactions in environmental decontamination. Front. Plant Sci. 2019, 10, 1519. [Google Scholar] [CrossRef]
- Fendrihan, S.; Pop, C. Biotechnological potential of plant associated microorganisms. Rom. Biotechnol. Lett. 2021, 26, 2700–2706. [Google Scholar] [CrossRef]
- Sivaranjani, M.; Srinivasan, R.; Aravindraja, C.; Karutha Pandian, S.; Veera Ravi, A. Inhibitory effect of α-mangostin on Acinetobacter baumannii biofilms—An in vitro study. Biofouling 2018, 34, 579–593. [Google Scholar] [CrossRef] [PubMed]
- Ramanathan, S.; Arunachalam, K.; Chandran, S.; Selvaraj, R.; Shunmugiah, K.; Arumugam, V. Biofilm inhibitory efficiency of phytol in combination with cefotaxime against nosocomial pathogen Acinetobacter baumannii. J. Appl. Microbiol. 2018, 125, 56–71. [Google Scholar] [CrossRef]
- Srinivasan, R.; Mohankumar, R.; Kannappan, A.; Karthick Raja, V.; Archunan, G.; Karutha Pandian, S.; Ruckmani, K.; Veera Ravi, A. Exploring the anti-quorum sensing and antibiofilm efficacy of phytol against Serratia marcescens associated acute pyelonephritis infection in Wistar rats. Front. Cell. Infect. Microbiol. 2017, 7, 498. [Google Scholar] [CrossRef] [Green Version]
- Srinivasan, R.; Devi, K.R.; Santhakumari, S.; Kannappan, A.; Chen, X.; Ravi, A.V.; Lin, X. Anti-quorum sensing and protective efficacies of naringin against Aeromonas hydrophila infection in Danio rerio. Front. Microbiol. 2020, 11, 3087. [Google Scholar] [CrossRef]
- Kannappan, A.; Balasubramaniam, B.; Ranjitha, R.; Srinivasan, R.; Packiavathy, I.A.S.V.; Balamurugan, K.; Pandian, S.K.; Ravi, A.V. In vitro and in vivo biofilm inhibitory efficacy of geraniol-cefotaxime combination against Staphylococcus spp. Food Chem. Toxicol. 2019, 125, 322–332. [Google Scholar] [CrossRef] [PubMed]
- Taylor, P.L.; Wright, G.D. Novel approaches to discovery of antibacterial agents. Anim. Health Res. Rev. 2008, 9, 237–246. [Google Scholar] [CrossRef] [PubMed]
- Fenical, W. Chemical studies of marine bacteria: Developing a new resource. Chem. Rev. 1993, 93, 1673–1683. [Google Scholar] [CrossRef]
- Aneiros, A.; Garateix, A. Bioactive peptides from marine sources: Pharmacological properties and isolation procedures. J. Chromatogr. B 2004, 803, 41–53. [Google Scholar] [CrossRef] [PubMed]
- Tziveleka, L.-A.; Vagias, C.; Roussis, V. Natural products with anti-HIV activity from marine organisms. Curr. Top. Med. Chem. 2003, 3, 1512–1535. [Google Scholar] [CrossRef] [PubMed]
- Gavriilidou, A.; Mackenzie, T.A.; Sánchez, P.; Tormo, J.R.; Ingham, C.; Smidt, H.; Sipkema, D. Bioactivity screening and gene-trait matching across marine sponge-associated bacteria. Mar. Drugs 2021, 19, 75. [Google Scholar] [CrossRef] [PubMed]
- Jaspars, M.; De Pascale, D.; Andersen, J.H.; Reyes, F.; Crawford, A.D.; Ianora, A. The marine biodiscovery pipeline and ocean medicines of tomorrow. J. Mar. Biol. Assoc. UK 2016, 96, 151–158. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.; Chen, J.; Hu, G.; Yu, J.; Zhu, X.; Lin, Y.; Chen, S.; Yuan, J. Statistical research on the bioactivity of new marine natural products discovered during the 28 years from 1985 to 2012. Mar. Drugs 2015, 13, 202–221. [Google Scholar] [CrossRef] [PubMed]
- Nalini, S.; Richard, D.S.; Riyaz, S.M.; Kavitha, G.; Inbakandan, D. Antibacterial macro molecules from marine organisms. Int. J. Biol. Macromol. 2018, 115, 696–710. [Google Scholar] [CrossRef]
- Abdelmohsen, U.R.; Balasubramanian, S.; Oelschlaeger, T.A.; Grkovic, T.; Pham, N.B.; Quinn, R.J.; Hentschel, U. Potential of marine natural products against drug-resistant fungal, viral, and parasitic infections. Lancet Infect. Dis. 2017, 17, e30–e41. [Google Scholar] [CrossRef]
- Fahmy, N.M.; Abdel-Tawab, A.M. Isolation and characterization of marine sponge–associated Streptomyces sp. NMF6 strain producing secondary metabolite (s) possessing antimicrobial, antioxidant, anticancer, and antiviral activities. J. Genet. Eng. Biotechnol. 2021, 19, 102. [Google Scholar] [CrossRef]
- Bhatnagar, I.; Kim, S.-K. Immense essence of excellence: Marine microbial bioactive compounds. Mar. Drugs 2010, 8, 2673–2701. [Google Scholar] [CrossRef] [Green Version]
- Laatsch, H. Marine bacterial metabolites. In Frontiers in Marine Biotechnology; Proksch, P., Müller, W.E.G., Eds.; Taylor & Francis: New York, NY, USA, 2006; Chapter 6. [Google Scholar]
- Piel, J. Metabolites from symbiotic bacteria. Nat. Prod. Rep. 2009, 26, 338–362. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, E.; Yan, L.; Boyd, K.G.; Wright, P.C.; Burgess, J.G. The symbiotic role of marine microbes on living surfaces. Hydrobiologia 2001, 461, 37–40. [Google Scholar] [CrossRef]
- Valliappan, K.; Sun, W.; Li, Z. Marine actinobacteria associated with marine organisms and their potentials in producing pharmaceutical natural products. Appl. Microbiol. Biotechnol. 2014, 98, 7365–7377. [Google Scholar] [CrossRef]
- Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2021, 38, 362–413. [Google Scholar] [CrossRef] [PubMed]
- Saurav, K.; Costantino, V.; Venturi, V.; Steindler, L. Quorum sensing inhibitors from the sea discovered using bacterial N-acyl-homoserine lactone-based biosensors. Mar. Drugs 2017, 15, 53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stincone, P.; Brandelli, A. Marine bacteria as source of antimicrobial compounds. Crit. Rev. Biotechnol. 2020, 40, 306–319. [Google Scholar] [CrossRef]
- Núñez-Montero, K.; Barrientos, L. Advances in Antarctic research for antimicrobial discovery: A comprehensive narrative review of bacteria from Antarctic environments as potential sources of novel antibiotic compounds against human pathogens and microorganisms of industrial importance. Antibiotics 2018, 7, 90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yasir, M. Analysis of bacterial communities and characterization of antimicrobial strains from cave microbiota. Braz. J. Microbiol. 2018, 49, 248–257. [Google Scholar] [CrossRef] [PubMed]
- Kuo, J.; Yang, Y.-T.; Lu, M.-C.; Wong, T.-Y.; Sung, P.-J.; Huang, Y.-S. Antimicrobial activity and diversity of bacteria associated with Taiwanese marine sponge Theonella swinhoei. Ann. Microbiol. 2019, 69, 253–265. [Google Scholar] [CrossRef]
- Graça, A.P.; Viana, F.; Bondoso, J.; Correia, M.I.; Gomes, L.; Humanes, M.; Reis, A.; Xavier, J.R.; Gaspar, H.; Lage, O.M. The antimicrobial activity of heterotrophic bacteria isolated from the marine sponge Erylus deficiens (Astrophorida, Geodiidae). Front. Microbiol. 2015, 6, 389. [Google Scholar]
- Albakosh, M.A.; Naidoo, R.K.; Kirby, B.; Bauer, R. Identification of epiphytic bacterial communities associated with the brown alga Splachnidium rugosum. J. Appl. Phycol. 2016, 28, 1891–1901. [Google Scholar] [CrossRef]
- Bull, A.T.; Stach, J.E. Marine actinobacteria: New opportunities for natural product search and discovery. Trends Microbiol. 2007, 15, 491–499. [Google Scholar] [CrossRef] [PubMed]
- Maldonado, L.A.; Stach, J.E.; Pathom-aree, W.; Ward, A.C.; Bull, A.T.; Goodfellow, M. Diversity of cultivable actinobacteria in geographically widespread marine sediments. Antonie Van Leeuwenhoek 2005, 87, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Stach, E.; Bull, A.T. Estimating and comparing the diversity of marine actinobacteria. Antonie Van Leeuwenhoek 2005, 87, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Jose, P.A.; Jha, B. Intertidal marine sediment harbours Actinobacteria with promising bioactive and biosynthetic potential. Sci. Rep. 2017, 7, 10041. [Google Scholar] [CrossRef]
- Montalvo, N.F.; Mohamed, N.M.; Enticknap, J.J.; Hill, R.T. Novel actinobacteria from marine sponges. Antonie Van Leeuwenhoek 2005, 87, 29–36. [Google Scholar] [CrossRef]
- Zhang, H.; Lee, Y.K.; Zhang, W.; Lee, H.K. Culturable actinobacteria from the marine sponge Hymeniacidon perleve: Isolation and phylogenetic diversity by 16S rRNA gene-RFLP analysis. Antonie Van Leeuwenhoek 2006, 90, 159–169. [Google Scholar] [CrossRef]
- Abdelmohsen, U.R.; Pimentel-Elardo, S.M.; Hanora, A.; Radwan, M.; Abou-El-Ela, S.H.; Ahmed, S.; Hentschel, U. Isolation, phylogenetic analysis and anti-infective activity screening of marine sponge-associated actinomycetes. Mar. Drugs 2010, 8, 399–412. [Google Scholar] [CrossRef]
- Xi, L.; Ruan, J.; Huang, Y. Diversity and biosynthetic potential of culturable actinomycetes associated with marine sponges in the China seas. Int. J. Mol. Sci. 2012, 13, 5917–5932. [Google Scholar] [CrossRef]
- Maldonado, L.A.; Fragoso-Yáñez, D.; Pérez-García, A.; Rosellón-Druker, J.; Quintana, E.T. Actinobacterial diversity from marine sediments collected in Mexico. Antonie Van Leeuwenhoek 2009, 95, 111–120. [Google Scholar] [CrossRef]
- Pesic, A.; Baumann, H.I.; Kleinschmidt, K.; Ensle, P.; Wiese, J.; Süssmuth, R.D.; Imhoff, J.F. Champacyclin, a new cyclic octapeptide from Streptomyces strain C42 isolated from the Baltic Sea. Mar. Drugs 2013, 11, 4834–4857. [Google Scholar] [CrossRef] [Green Version]
- Contreras-Castro, L.; Martinez-Garcia, S.; Cancino-Diaz, J.C.; Maldonado, L.A.; Hernandez-Guerrero, C.J.; Martínez-Díaz, S.F.; Gonzalez-Acosta, B.; Quintana, E.T. Marine sediment recovered Salinispora sp. inhibits the growth of emerging bacterial pathogens and other multi-drug-resistant bacteria. Pol. J. Microbiol. 2020, 69, 321. [Google Scholar] [CrossRef] [PubMed]
- Yücel, S.; Yamaç, M. Selection of Streptomyces isolates from Turkish karstic caves against antibiotic resistant microorganisms. Pak. J. Pharm. Sci 2010, 23, 1–6. [Google Scholar]
- Kishimoto, S.; Nishimura, S.; Hattori, A.; Tsujimoto, M.; Hatano, M.; Igarashi, M.; Kakeya, H. Chlorocatechelins A and B from Streptomyces sp.: New siderophores containing chlorinated catecholate groups and an acylguanidine structure. Org. Lett. 2014, 16, 6108–6111. [Google Scholar] [CrossRef] [PubMed]
- Ser, H.-L.; Ab Mutalib, N.-S.; Yin, W.-F.; Chan, K.-G.; Goh, B.-H.; Lee, L.-H. Evaluation of antioxidative and cytotoxic activities of Streptomyces pluripotens MUSC 137 isolated from mangrove soil in Malaysia. Front. Microbiol. 2015, 6, 1398. [Google Scholar] [CrossRef] [Green Version]
- Lee, L.-H.; Zainal, N.; Azman, A.-S.; Eng, S.-K.; Ab Mutalib, N.-S.; Yin, W.-F.; Chan, K.-G. Streptomyces pluripotens sp. nov., a bacteriocin-producing streptomycete that inhibits meticillin-resistant Staphylococcus aureus. Int. J. Syst. Evol. Microbiol. 2014, 64, 3297–3306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sajid, I.; Shaaban, K.A.; Hasnain, S. Identification, isolation and optimization of antifungal metabolites from the Streptomyces Malachitofuscus ctf9. Braz. J. Microbiol. 2011, 42, 592–604. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernández-Gomez, B.; Richter, M.; Schüler, M.; Pinhassi, J.; Acinas, S.G.; González, J.M.; Pedros-Alio, C. Ecology of marine Bacteroidetes: A comparative genomics approach. ISME J. 2013, 7, 1026–1037. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ludwig, W.; Euzéby, J.; Whitman, W. Taxonomic Outline of the Phyla Bacteroidetes, Spirochaetes, Tenericutes (Mollicutes), Acidobacteria, Fibrobacteres, Fusobacteria, Dictyoglomi, Gemmatimonadetes, Lentisphaerae, Verrucomicrobia, Chlamydiae, and Planctomycetes. In Bergey’s Manual of Systematic Bacteriology, 2nd ed.; Staley, J.T., Hedlund, B., Paster, B.J., Ward, N., Ludwig, W., Whitman, W.B., Eds.; Springer: Berlin, Germany, 2009; Volume 4. [Google Scholar]
- Belizário, J.E.; Napolitano, M. Human microbiomes and their roles in dysbiosis, common diseases, and novel therapeutic approaches. Front. Microbiol. 2015, 6, 1050. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gavriilidou, A.; Gutleben, J.; Versluis, D.; Forgiarini, F.; Van Passel, M.W.; Ingham, C.J.; Smidt, H.; Sipkema, D. Comparative genomic analysis of Flavobacteriaceae: Insights into carbohydrate metabolism, gliding motility and secondary metabolite biosynthesis. BMC Genom. 2020, 21, 569. [Google Scholar] [CrossRef]
- León, J.; Liza, L.; Soto, I.; Torres, M.; Orosco, A. Marine bacteria producing antibacterial compounds isolated from inter-tidal invertebrates. Rev. Peru. Med. Exp. Salud Publ. 2010, 27, 215–221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zia, M.A.; Ishaq, M.; Ahmed, I.; Iqbal, H.M.N. Studies on antifungal and antibacterial activity of glucose aero dehydrogenase. World Appl. Sci. J. 2012, 18, 166–170. [Google Scholar]
- Tsugawa, W.; Horiuchi, S.; Tanaka, M.; Wake, H.; Sode, K. Purification of a marine bacterial glucose dehydrogenase from Cytophaga marinoflava and its application for measurement of 1, 5-anhydro-D-glucitol. Appl. Biochem. Biotechnol. 1996, 56, 301–310. [Google Scholar] [CrossRef] [PubMed]
- Singh, J.S.; Kumar, A.; Rai, A.N.; Singh, D.P. Cyanobacteria: A precious bio-resource in agriculture, ecosystem, and environmental sustainability. Front. Microbiol. 2016, 7, 529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, L.T. Bioactive natural products from marine cyanobacteria for drug discovery. Phytochemistry 2007, 68, 954–979. [Google Scholar] [CrossRef] [PubMed]
- Cardellina, J.H.; Moore, G.E.; Bradley, S. Richard E. Moore (1933–2007); ACS Publications: Washington, DC, USA, 2010. [Google Scholar]
- Tidgewell, K.; Clark, B.; Gerwick, W. Comprehensive Natural Products Chemistry; Pergamon Press: Oxford, UK, 2010. [Google Scholar]
- Nagarajan, M.; Maruthanayagam, V.; Sundararaman, M. A review of pharmacological and toxicological potentials of marine cyanobacterial metabolites. J. Appl. Toxicol. 2012, 32, 153–185. [Google Scholar] [CrossRef]
- Mo, S.; Krunic, A.; Chlipala, G.; Orjala, J. Antimicrobial ambiguine isonitriles from the cyanobacterium Fischerella ambigua. J. Nat. Prod. 2009, 72, 894–899. [Google Scholar] [CrossRef] [Green Version]
- Brumley, D.; Spencer, K.A.; Gunasekera, S.P.; Sauvage, T.; Biggs, J.; Paul, V.J.; Luesch, H. Isolation and characterization of anaephenes A–C, alkylphenols from a filamentous cyanobacterium (Hormoscilla sp., oscillatoriales). J. Nat. Prod. 2018, 81, 2716–2721. [Google Scholar] [CrossRef] [PubMed]
- Reshef, V.; Mizrachi, E.; Maretzki, T.; Silberstein, C.; Loya, S.; Hizi, A.; Carmeli, S. New acylated sulfoglycolipids and digalactolipids and related known glycolipids from cyanobacteria with a potential to inhibit the reverse transcriptase of HIV-1. J. Nat. Prod. 1997, 60, 1251–1260. [Google Scholar] [CrossRef]
- Gordon, D.M.; Danishefsky, S.J. Synthesis of a cyanobacterial sulfolipid: Confirmation of its structure, stereochemistry and anti-HIV-1 activity. J. Am. Chem. Soc. 1992, 114, 659–663. [Google Scholar] [CrossRef]
- Loya, S.; Reshef, V.; Mizrachi, E.; Silberstein, C.; Rachamim, Y.; Carmeli, S.; Hizi, A. The inhibition of the reverse transcriptase of HIV-1 by the natural sulfoglycolipids from cyanobacteria: Contribution of different moieties to their high potency. J. Nat. Prod. 1998, 61, 891–895. [Google Scholar] [CrossRef]
- Gustafson, K.R.; Cardellina, J.H.; Fuller, R.W.; Weislow, O.S.; Kiser, R.F.; Snader, K.M.; Patterson, G.M.; Boyd, M.R. AIDS-antiviral sulfolipids from cyanobacteria (blue-green algae). JNCI J. Natl. Cancer Inst. 1989, 81, 1254–1258. [Google Scholar] [CrossRef] [PubMed]
- Ayehunie, S.; Belay, A.; Baba, T.W.; Ruprecht, R.M. Inhibition of HIV-1 replication by an aqueous extract of Spirulina platensis (Arthrospira platensis). J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 1998, 18, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, T.; Hayashi, K.; Maeda, M.; Kojima, I. Calcium spirulan, an inhibitor of enveloped virus replication, from a blue-green alga Spirulina platensis. J. Nat. Prod. 1996, 59, 83–87. [Google Scholar] [CrossRef] [PubMed]
- Boyd, M.R.; Gustafson, K.R.; McMahon, J.B.; Shoemaker, R.H.; O’Keefe, B.R.; Mori, T.; Gulakowski, R.J.; Wu, L.; Rivera, M.I.; Laurencot, C.M. Discovery of cyanovirin-N, a novel human immunodeficiency virus-inactivating protein that binds viral surface envelope glycoprotein gp120: Potential applications to microbicide development. Antimicrob. Agents Chemother. 1997, 41, 1521–1530. [Google Scholar] [CrossRef] [Green Version]
- Ntougias, S.; Russell, N.J. Bacillus sp. WW3-SN6, a novel facultatively alkaliphilic bacterium isolated from the washwaters of edible olives. Extremophiles 2000, 4, 201–208. [Google Scholar] [CrossRef]
- Rossi, M.; Ciaramella, M.; Cannio, R.; Pisani, F.M.; Moracci, M.; Bartolucci, S. Extremophiles 2002. J. Bacteriol. 2003, 185, 3683–3689. [Google Scholar] [CrossRef] [Green Version]
- Nair, A.V.; Praveen, N.; Joseph, N.; Leo, A.M.; Vijayan, K. Isolation and characterization of a novel antimicrobial oxatetracyclo ketone from Bacillus stercoris MBTDCMFRI Ba37 isolated from the tropical estuarine habitats of Cochin. Mol. Biol. Rep. 2021, 48, 1299–1310. [Google Scholar] [CrossRef]
- Vairagkar, U.; Mirza, Y. Antagonistic activity of antimicrobial metabolites produced from seaweed-associated Bacillus amyloliquefaciens MTCC 10456 Against Malassezia spp. Probiot. Antimicrob. Proteins 2021, 13, 1228–1237. [Google Scholar] [CrossRef]
- Wang, W.; Park, K.-H.; Lee, J.; Oh, E.; Park, C.; Kang, E.; Lee, J.; Kang, H. A new thiopeptide antibiotic, Micrococcin P3, from a marine-derived strain of the bacterium Bacillus stratosphericus. Molecules 2020, 25, 4383. [Google Scholar] [CrossRef]
- Fuerst, J.A.; Sagulenko, E. Beyond the bacterium: Planctomycetes challenge our concepts of microbial structure and function. Nat. Rev. Microbiol. 2011, 9, 403–413. [Google Scholar] [CrossRef] [PubMed]
- Sandargo, B.; Jeske, O.; Boedeker, C.; Wiegand, S.; Wennrich, J.-P.; Kallscheuer, N.; Jogler, M.; Rohde, M.; Jogler, C.; Surup, F. Stieleriacines, N-acyl dehydrotyrosines from the marine planctomycete Stieleria neptunia sp. nov. Front. Microbiol. 2020, 11, 1408. [Google Scholar] [CrossRef]
- Jeske, O.; Jogler, M.; Petersen, J.; Sikorski, J.; Jogler, C. From genome mining to phenotypic microarrays: Planctomycetes as source for novel bioactive molecules. Antonie Van Leeuwenhoek 2013, 104, 551–567. [Google Scholar] [CrossRef] [PubMed]
- Jeske, O.; Surup, F.; Ketteniß, M.; Rast, P.; Förster, B.; Jogler, M.; Wink, J.; Jogler, C. Developing techniques for the utilization of Planctomycetes as producers of bioactive molecules. Front. Microbiol. 2016, 7, 1242. [Google Scholar] [CrossRef] [Green Version]
- Kallscheuer, N.; Jeske, O.; Sandargo, B.; Boedeker, C.; Wiegand, S.; Bartling, P.; Jogler, M.; Rohde, M.; Petersen, J.; Medema, M.H. The planctomycete Stieleria maiorica Mal15 T employs stieleriacines to alter the species composition in marine biofilms. Commun. Biol. 2020, 3, 303. [Google Scholar] [CrossRef]
- Liao, L.; Xu, X.-W.; Wang, C.-S.; Zhang, D.-S.; Wu, M. Bacterial and archaeal communities in the surface sediment from the northern slope of the South China Sea. J. Zhejiang Univ. Sci. B 2009, 10, 890–901. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kai, W.; Peisheng, Y.; Rui, M.; Wenwen, J.; Zongze, S. Diversity of culturable bacteria in deep-sea water from the South Atlantic Ocean. Bioengineered 2017, 8, 572–584. [Google Scholar] [CrossRef] [Green Version]
- Viju, N.; Punitha, S.M.J.; Satheesh, S. An analysis of biosynthesis gene clusters and bioactivity of marine bacterial symbionts. Curr. Microbiol. 2021, 78, 2522–2533. [Google Scholar] [CrossRef]
- Bowman, J.P. Bioactive compound synthetic capacity and ecological significance of marine bacterial genus Pseudoalteromonas. Mar. Drugs 2007, 5, 220–241. [Google Scholar] [CrossRef] [PubMed]
- Dat, T.T.H.; Cuc, N.T.K.; Cuong, P.V.; Smidt, H.; Sipkema, D. Diversity and antimicrobial activity of Vietnamese sponge-associated bacteria. Mar. Drugs 2021, 19, 353. [Google Scholar] [CrossRef] [PubMed]
- Fenical, W.; Jensen, P.R. Developing a new resource for drug discovery: Marine actinomycete bacteria. Nat. Chem. Biol. 2006, 2, 666–673. [Google Scholar] [CrossRef]
- Habbu, P.; Warad, V.; Shastri, R.; Madagundi, S.; Kulkarni, V.H. Antimicrobial metabolites from marine microorganisms. Chin. J. Nat. Med. 2016, 14, 101–116. [Google Scholar] [CrossRef]
- You, J.; Cao, L.; Liu, G.; Zhou, S.; Tan, H.; Lin, Y. Isolation and characterization of actinomycetes antagonistic to pathogenic Vibrio spp. from nearshore marine sediments. World J. Microbiol. Biotechnol. 2005, 21, 679–682. [Google Scholar] [CrossRef]
- Asolkar, R.N.; Maskey, R.P.; Helmke, E.; Laatsch, H. Chalcomycin B, a new macrolide antibiotic from the marine isolate Streptomyces sp. B7064. J. Antibiot. 2002, 55, 893–898. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shao, M.; Ma, J.; Li, Q.; Ju, J. Identification of the anti-infective aborycin biosynthetic gene cluster from deep-sea-derived Streptomyces sp. SCSIO ZS0098 enables production in a heterologous host. Mar. Drugs 2019, 17, 127. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Li, B.; Qin, Y.; Karthik, L.; Zhu, G.; Hou, C.; Jiang, L.; Liu, M.; Ye, X.; Liu, M. A new abyssomicin polyketide with anti-influenza A virus activity from a marine-derived Verrucosispora sp. MS100137. Appl. Microbiol. Biotechnol. 2020, 104, 1533–1543. [Google Scholar] [CrossRef] [PubMed]
- Grossart, H.-P.; Thorwest, M.; Plitzko, I.; Brinkhoff, T.; Simon, M.; Zeeck, A. Production of a blue pigment (Glaukothalin) by marine Rheinheimera spp. Int. J. Microbiol. 2009, 2009, 701735. [Google Scholar] [CrossRef] [Green Version]
- Soliev, A.B.; Hosokawa, K.; Enomoto, K. Bioactive pigments from marine bacteria: Applications and physiological roles. Evid.-Based Complement. Altern. Med. 2011, 2011, 670349. [Google Scholar] [CrossRef] [PubMed]
- Boger, D.L.; Patel, M. Total synthesis of prodigiosin. Tetrahedron Lett. 1987, 28, 2499–2502. [Google Scholar] [CrossRef]
- Gandhi, N.; Patell, J.; Gandhi, J.; De Souza, N.; Kohl, H. Prodigiosin metabolites of a marine Pseudomonas species. Mar. Biol. 1976, 34, 223–227. [Google Scholar] [CrossRef]
- Gauthier, M. Alteromonas rubra sp. nov., a new marine antibiotic-producing bacterium. Int. J. Syst. Evol. Microbiol. 1976, 26, 459–466. [Google Scholar] [CrossRef] [Green Version]
- Lewis, S.; Corpe, W. Prodigiosin-producing bacteria from marine sources. Appl. Microbiol. 1964, 12, 13–17. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, T.; Kurachi, M.; Kato, Y.; Yamaguchi, K.; Oda, T. Characterization of bacterium isolated from the sediment at coastal area of Omura Bay in Japan and several biological activities of pigment produced by this isolate. Microbiol. Immunol. 2005, 49, 407–415. [Google Scholar] [CrossRef]
- Le, T.C.; Yang, I.; Yoon, Y.J.; Nam, S.-J.; Fenical, W. Ansalactams B–D illustrate further biosynthetic plasticity within the ansamycin pathway. Org. Lett. 2016, 18, 2256–2259. [Google Scholar] [CrossRef] [PubMed]
- Hong, W.Y.; Jun, Y.S.; Yu, S.J.; Mei, Z.L.; Hong, L. Recent advances in study on cyclic peptides from marine sponges. Chin. J. Org. Chem. 2001, 21, 16–24. [Google Scholar]
- Xu, X.; Han, J.; Lin, R.; Polyak, S.W.; Song, F. Two new piperazine-triones from a marine-derived Streptomycetes sp. strain SMS636. Mar. Drugs 2019, 17, 186. [Google Scholar] [CrossRef] [Green Version]
- Engelhardt, K.; Degnes, K.F.; Kemmler, M.; Bredholt, H.; Fjærvik, E.; Klinkenberg, G.; Sletta, H.; Ellingsen, T.E.; Zotchev, S.B. Production of a new thiopeptide antibiotic, TP-1161, by a marine Nocardiopsis species. Appl. Environ. Microbiol. 2010, 76, 4969–4976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, P.; Maloney, K.N.; Nam, S.-J.; Haste, N.M.; Raju, R.; Aalbersberg, W.; Jensen, P.R.; Nizet, V.; Hensler, M.E.; Fenical, W. Fijimycins A–C, three antibacterial etamycin-class depsipeptides from a marine-derived Streptomyces sp. Bioorg. Med. Chem. 2011, 19, 6557–6562. [Google Scholar] [CrossRef] [Green Version]
- Bertrand, S.; Bohni, N.; Schnee, S.; Schumpp, O.; Gindro, K.; Wolfender, J.-L. Metabolite induction via microorganism co-culture: A potential way to enhance chemical diversity for drug discovery. Biotechnol. Adv. 2014, 32, 1180–1204. [Google Scholar] [CrossRef]
- Anjum, K.; Sadiq, I.; Chen, L.; Kaleem, S.; Li, X.-C.; Zhang, Z.; Lian, X.-Y. Novel antifungal janthinopolyenemycins A and B from a co-culture of marine-associated Janthinobacterium spp. ZZ145 and ZZ148. Tetrahedron Lett. 2018, 59, 3490–3494. [Google Scholar] [CrossRef]
- Cho, J.Y.; Kim, M.S. Induction of antifouling diterpene production by Streptomyces cinnabarinus PK209 in co-culture with marine-derived Alteromonas sp. KNS-16. Biosci. Biotechnol. Biochem. 2012, 76, 1849–1854. [Google Scholar] [CrossRef] [Green Version]
- Oku, N.; Kawabata, K.; Adachi, K.; Katsuta, A.; Shizuri, Y. Unnarmicins A and C, new antibacterial depsipeptides produced by marine bacterium Photobacterium sp. MBIC06485. J. Antibiot. 2008, 61, 11–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Braña, A.F.; Sarmiento-Vizcaíno, A.; Pérez-Victoria, I.; Otero, L.; Fernández, J.; Palacios, J.J.; Martín, J.S.; De la Cruz, M.; Díaz, C.; Vicente, F. Branimycins B and C, antibiotics produced by the abyssal actinobacterium Pseudonocardia carboxydivorans M-227. J. Nat. Prod. 2017, 80, 569–573. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.-S.; Eom, S.-H.; Jeong, S.-Y.; Shin, H.J.; Je, J.-Y.; Lee, E.-W.; Chung, Y.-H.; Kim, Y.-M.; Kang, C.-K.; Lee, M.-S. Anti-methicillin-resistant Staphylococcus aureus (MRSA) substance from the marine bacterium Pseudomonas sp. UJ-6. Environ. Toxicol. Pharmacol. 2013, 35, 171–177. [Google Scholar] [CrossRef] [PubMed]
- Furumai, T.; Takagi, K.; Igarashi, Y.; Saito, N.; Oki, T. Arisostatins A and B, new members of tetrocarcin class of antibiotics from Micromonospora sp. TP-A0316 I. Taxonomy, fermentation, isolation and biological properties. J. Antibiot. 2000, 53, 227–232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arena, A.; Maugeri, T.L.; Pavone, B.; Iannello, D.; Gugliandolo, C.; Bisignano, G. Antiviral and immunoregulatory effect of a novel exopolysaccharide from a marine thermotolerant Bacillus licheniformis. Int. Immunopharmacol. 2006, 6, 8–13. [Google Scholar] [CrossRef]
- Arena, A.; Gugliandolo, C.; Stassi, G.; Pavone, B.; Iannello, D.; Bisignano, G.; Maugeri, T.L. An exopolysaccharide produced by Geobacillus thermodenitrificans strain B3-72: Antiviral activity on immunocompetent cells. Immunol. Lett. 2009, 123, 132–137. [Google Scholar] [CrossRef]
- Igarashi, M.; Sawa, R.; Umekita, M.; Hatano, M.; Arisaka, R.; Hayashi, C.; Ishizaki, Y.; Suzuki, M.; Kato, C. Sealutomicins, new enediyne antibiotics from the deep-sea actinomycete Nonomuraea sp. MM565M-173N2. J. Antibiot. 2021, 74, 291–299. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, I.; Girão, M.; Alexandrino, D.A.; Ribeiro, T.; Santos, C.; Pereira, F.; Mucha, A.P.; Urbatzka, R.; Leão, P.N.; Carvalho, M.F. Diversity and bioactive potential of actinobacteria isolated from a coastal marine sediment in northern Portugal. Microorganisms 2020, 8, 1691. [Google Scholar] [CrossRef] [PubMed]
- Sangkanu, S.; Rukachaisirikul, V.; Suriyachadkun, C.; Phongpaichit, S. Antifungal activity of marine-derived actinomycetes against Talaromyces marneffei. J. Appl. Microbiol. 2021, 130, 1508–1522. [Google Scholar] [CrossRef] [PubMed]
- Yi, W.; Qin, L.; Lian, X.-Y.; Zhang, Z. New antifungal metabolites from the mariana trench sediment-associated actinomycete Streptomyces sp. SY1965. Mar. Drugs 2020, 18, 385. [Google Scholar] [CrossRef]
- Ma, Z.; Zhang, S.; Zhang, S.; Wu, G.; Shao, Y.; Mi, Q.; Liang, J.; Sun, K.; Hu, J. Isolation and characterization of a new cyclic lipopeptide surfactin from a marine-derived Bacillus velezensis SH-B74. J. Antibiot. 2020, 73, 863–867. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Ding, H.; Yu, Y.; Chen, B. A cold-adapted chitinase-producing bacterium from Antarctica and its potential in biocontrol of plant pathogenic fungi. Mar. Drugs 2019, 17, 695. [Google Scholar] [CrossRef] [Green Version]
- Choi, A.; Nam, Y.H.; Baek, K.; Chung, E.J. Brevibacillus antibioticus sp. nov., with a broad range of antibacterial activity, isolated from soil in the Nakdong River. J. Microbiol. 2019, 57, 991–996. [Google Scholar] [CrossRef] [PubMed]
- Siddharth, S. Isolation and characterization of bioactive compounds with antibacterial, antioxidant and enzyme inhibitory activities from marine-derived rare actinobacteria, Nocardiopsis sp. SCA21. Microb. Pathog. 2019, 137, 103775. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, A.B.; Pinto, F.C.; Silveira, E.R.; Costa-Lotufo, L.V.; Costa, W.S.; Ayala, A.P.; Canuto, K.M.; Barros, A.B.; Araújo, A.J.; Marinho Filho, J.D.B. 4-Hydroxy-pyran-2-one and 3-hydroxy-N-methyl-2-oxindole derivatives of Salinispora arenicola from Brazilian marine sediments. Fitoterapia 2019, 138, 104357. [Google Scholar] [CrossRef]
- Almalki, M.A. Isolation and characterization of polyketide drug molecule from Streptomyces species with antimicrobial activity against clinical pathogens. J. Infect. Public Health 2020, 13, 125–130. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, A.B.; Silveira, E.R.; Wilke, D.V.; Ferreira, E.G.; Costa-Lotufo, L.V.; Torres, M.C.M.; Ayala, A.P.; Costa, W.S.; Canuto, K.M.; De Araujo-Nobre, A.R. Antibacterial Salinaphthoquinones from a strain of the bacterium Salinispora arenicola recovered from the marine sediments of St. Peter and St. Paul Archipelago, Brazil. J. Nat. Prod. 2019, 82, 1831–1838. [Google Scholar] [CrossRef]
- Ryu, M.-J.; Hwang, S.; Kim, S.; Yang, I.; Oh, D.-C.; Nam, S.-J.; Fenical, W. Meroindenon and merochlorins E and F, antibacterial meroterpenoids from a marine-derived sediment bacterium of the genus Streptomyces. Org. Lett. 2019, 21, 5779–5783. [Google Scholar] [CrossRef]
- Sproule, A.; Correa, H.; Decken, A.; Haltli, B.; Berrué, F.; Overy, D.P.; Kerr, R.G. Terrosamycins A and B, bioactive polyether ionophores from Streptomyces sp. RKND004 from Prince Edward Island sediment. Mar. Drugs 2019, 17, 347. [Google Scholar] [CrossRef] [Green Version]
- Gozari, M.; Zaheri, A.; Jahromi, S.T.; Gozari, M.; Karimzadeh, R. Screening and characterization of marine actinomycetes from the northern Oman Sea sediments for cytotoxic and antimicrobial activity. Int. Microbiol. 2019, 22, 521–530. [Google Scholar] [CrossRef]
- De Oliveira, J.A.M.; Williams, D.E.; Andersen, R.J.; Sarragiotto, M.H.; Baldoqui, D.C. Mycenolide A, new butenolide from a marine sediment-derived bacterium Streptomyces sp. 4054. Nat. Prod. Res. 2020, 34, 2986–2989. [Google Scholar] [CrossRef]
- Zhang, S.; Gui, C.; Shao, M.; Kumar, P.S.; Huang, H.; Ju, J. Antimicrobial tunicamycin derivatives from the deep sea-derived Streptomyces xinghaiensis SCSIO S15077. Nat. Prod. Res. 2020, 34, 1499–1504. [Google Scholar] [CrossRef] [PubMed]
- Galaviz-Silva, L.; Iracheta-Villarreal, J.M.; Molina-Garza, Z.J. Bacillus and Virgibacillus strains isolated from three Mexican coasts antagonize Staphylococcus aureus and Vibrio parahaemolyticus. FEMS Microbiol. Lett. 2018, 365, fny202. [Google Scholar] [CrossRef] [PubMed]
- Priyanka, S.; Jayashree, M.; Shivani, R.; Anwesha, S.; Rao, K.B. Characterisation and identification of antibacterial compound from marine actinobacteria: In vitro and in silico analysis. J. Infect. Public Health 2019, 12, 83–89. [Google Scholar] [CrossRef]
- Quintero, M.; Velásquez, A.; Jutinico, L.; Jiménez-Vergara, E.; Blandón, L.; Martinez, K.; Lee, H.; Gómez-León, J. Bioprospecting from marine coastal sediments of Colombian Caribbean: Screening and study of antimicrobial activity. J. Appl. Microbiol. 2018, 125, 753–765. [Google Scholar] [CrossRef]
- Hamed, A.; Abdel-Razek, A.S.; Frese, M.; Wibberg, D.; El-Haddad, A.F.; Ibrahim, T.M.; Kalinowski, J.; Sewald, N.; Shaaban, M. N-Acetylborrelidin B: A new bioactive metabolite from Streptomyces mutabilis sp. MII. Z. Nat. C 2018, 73, 49–57. [Google Scholar] [CrossRef]
- Sangkanu, S.; Rukachaisirikul, V.; Suriyachadkun, C.; Phongpaichit, S. Evaluation of antibacterial potential of mangrove sediment-derived actinomycetes. Microb. Pathog. 2017, 112, 303–312. [Google Scholar] [CrossRef] [PubMed]
- Arumugam, T.; Kumar, P.S.; Kameshwar, R.; Prapanchana, K. Screening of novel actinobacteria and characterization of the potential isolates from mangrove sediment of south coastal India. Microb. Pathog. 2017, 107, 225–233. [Google Scholar] [CrossRef] [PubMed]
- Kurata, A.; Yamaura, Y.; Tanaka, T.; Kato, C.; Nakasone, K.; Kishimoto, N. Antifungal peptidic compound from the deep-sea bacterium Aneurinibacillus sp. YR247. World J. Microbiol. Biotechnol. 2017, 33, 73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalinovskaya, N.; Romanenko, L.; Kalinovsky, A. Antibacterial low-molecular-weight compounds produced by the marine bacterium Rheinheimera japonica KMM 9513. Antonie Van Leeuwenhoek 2017, 110, 719–726. [Google Scholar] [CrossRef]
- Anand, B.G.; Thomas, C.N.; Prakash, S. In vitro cytotoxicity and antimicrobial activity of Talaromyces flavus SP5 inhabited in the marine sediment of Southern Coast of India. Chin. J. Nat. Med. 2016, 14, 913–921. [Google Scholar] [CrossRef]
- Baskaran, R.; Mohan, P.; Sivakumar, K.; Kumar, A. Antimicrobial activity and phylogenetic analysis of Streptomyces parvulus DOSMB-D105 isolated from the mangrove sediments of Andaman Islands. Acta Microbiol. Immunol. Hungar. 2016, 63, 27–46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romanenko, L.; Tanaka, N.; Svetashev, V.; Kalinovskaya, N.; Mikhailov, V. Rheinheimera japonica sp. nov., a novel bacterium with antimicrobial activity from seashore sediments of the Sea of Japan. Arch. Microbiol. 2015, 197, 613–620. [Google Scholar] [CrossRef]
- Raju, R.; Khalil, Z.G.; Piggott, A.M.; Blumenthal, A.; Gardiner, D.L.; Skinner-Adams, T.S.; Capon, R.J. Mollemycin A: An antimalarial and antibacterial glyco-hexadepsipeptide-polyketide from an Australian marine-derived Streptomyces sp. (CMB-M0244). Org. Lett. 2014, 16, 1716–1719. [Google Scholar] [CrossRef]
- Chopra, L.; Singh, G.; Choudhary, V.; Sahoo, D.K. Sonorensin: An antimicrobial peptide, belonging to the heterocycloanthracin subfamily of bacteriocins, from a new marine isolate, Bacillus sonorensis MT93. Appl. Environ. Microbiol. 2014, 80, 2981–2990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sletta, H.; Degnes, K.F.; Herfindal, L.; Klinkenberg, G.; Fjærvik, E.; Zahlsen, K.; Brunsvik, A.; Nygaard, G.; Aachmann, F.L.; Ellingsen, T.E. Anti-microbial and cytotoxic 1, 6-dihydroxyphenazine-5, 10-dioxide (iodinin) produced by Streptosporangium sp. DSM 45942 isolated from the fjord sediment. Appl. Microbiol. Biotechnol. 2014, 98, 603–610. [Google Scholar] [CrossRef] [PubMed]
- Pan, H.-Q.; Zhang, S.-Y.; Wang, N.; Li, Z.-L.; Hua, H.-M.; Hu, J.-C.; Wang, S.-J. New spirotetronate antibiotics, lobophorins H and I, from a South China Sea-derived Streptomyces sp. 12A35. Mar. Drugs 2013, 11, 3891–3901. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sato, S.; Iwata, F.; Yamada, S.; Katayama, M. Neomaclafungins A–I: Oligomycin-class macrolides from a marine-derived actinomycete. J. Nat. Prod. 2012, 75, 1974–1982. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Tian, X.; Niu, S.; Zhang, W.; Chen, Y.; Zhang, H.; Yang, X.; Zhang, W.; Li, W.; Zhang, S. Pseudonocardians A–C, new diazaanthraquinone derivatives from a deap-sea actinomycete Pseudonocardia sp. SCSIO 01299. Mar. Drugs 2011, 9, 1428–1439. [Google Scholar] [CrossRef] [Green Version]
- Hohmann, C.; Schneider, K.; Bruntner, C.; Irran, E.; Nicholson, G.; Bull, A.T.; Jones, A.L.; Brown, R.; Stach, J.E.; Goodfellow, M. Caboxamycin, a new antibiotic of the benzoxazole family produced by the deep-sea strain Streptomyces sp. NTK 937. J. Antibiot. 2009, 62, 99–104. [Google Scholar] [CrossRef] [Green Version]
- Ren, Z.-Z.; Zheng, Y.; Sun, M.; Liu, J.-Z.; Wang, Y.-J. Purification and properties of an antimicrobial substance from marine Brevibacillus laterosporus Lh-1. Acta Microbiol. Sin. 2007, 47, 997–1001. [Google Scholar]
- Schumacher, R.W.; Talmage, S.C.; Miller, S.A.; Sarris, K.E.; Davidson, B.S.; Goldberg, A. Isolation and structure determination of an antimicrobial ester from a marine sediment-derived bacterium. J. Nat. Prod. 2003, 66, 1291–1293. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Song, Y.; Li, X.; Wang, X.; Ling, C.; Qin, X.; Zhou, Z.; Li, Q.; Wei, X.; Ju, J. Abyssomicin monomers and dimers from the marine-derived Streptomyces koyangensis SCSIO 5802. J. Nat. Prod. 2018, 81, 1892–1898. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-H.; Ha, T.-K.-Q.; Oh, W.K.; Shin, J.; Oh, D.-C. Antiviral indolosesquiterpenoid xiamycins C–E from a halophilic actinomycete. J. Nat. Prod. 2016, 79, 51–58. [Google Scholar] [CrossRef]
- Uzair, B.; Bano, A.; Niazi, M.B.K.; Khan, F.; Mujtaba, G. In vitro antifungal activity of 9, 10-dihydrophenanthrene-2-carboxylic acid isolated from a marine bacterium: Pseudomonas putida. Pak. J. Pharm. Sci. 2018, 31, 2733–2736. [Google Scholar]
- Harounabadi, S.; Shamsabad, P.E.; Mostafavi, S.K.S.; Meybodi, S.M. Isolation and partial characterization of bacteria with potential antimicrobial activity from the Caspian Sea. Russ. J. Mar. Biol. 2016, 42, 36–41. [Google Scholar] [CrossRef]
- Sannino, F.; Parrilli, E.; Apuzzo, G.A.; De Pascale, D.; Tedesco, P.; Maida, I.; Perrin, E.; Fondi, M.; Fani, R.; Marino, G. Pseudoalteromonas haloplanktis produces methylamine, a volatile compound active against Burkholderia cepacia complex strains. New Biotechnol. 2017, 35, 13–18. [Google Scholar] [CrossRef]
- Jayme, M.M.A.; Castro, R.O.; Silva, C.A.M.; Silva, M.M.; Do Carmo, F.L.; De Araujo, F.V. Evaluation of the biotechnological potential of bacterioplankton from Niterói coast, RJ. Compt. Rendus Biol. 2017, 340, 324–329. [Google Scholar] [CrossRef]
- Taylor, M.W.; Radax, R.; Steger, D.; Wagner, M. Sponge-associated microorganisms: Evolution, ecology, and biotechnological potential. Microbiol. Mol. Biol. Rev. 2007, 71, 295–347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sagar, S.; Kaur, M.; Minneman, K.P. Antiviral lead compounds from marine sponges. Mar. Drugs 2010, 8, 2619–2638. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wagner-Döbler, I.; Beil, W.; Lang, S.; Meiners, M.; Laatsch, H. Integrated approach to explore the potential of marine microorganisms for the production of bioactive metabolites. Tools Appl. Biochem. Eng. Sci. 2002, 74, 207–238. [Google Scholar]
- Thoms, C.; Schupp, P. Biotechnological Potential of Marine Sponges and Their Associated Bacteria as Producers of New Pharmaceuticals (Part II); Walter de Gruyter: Berlin, Germany, 2005. [Google Scholar]
- Bibi, F.; Naseer, M.I.; Azhar, E.I. Assessing the diversity of bacterial communities from marine sponges and their bioactive compounds. Saudi J. Biol. Sci. 2021, 28, 2747–2754. [Google Scholar] [CrossRef]
- Altuğ, G.; Türetken, P.S.Ç.; Kalkan, S.; Topaloğlu, B. The distribution and antibacterial activity of marine sponge-associated bacteria in the Aegean Sea and the Sea of Marmara, Turkey. Curr. Microbiol. 2021, 78, 2275–2290. [Google Scholar] [CrossRef] [PubMed]
- Abdelmohsen, U.R.; Bayer, K.; Hentschel, U. Diversity, abundance and natural products of marine sponge-associated actinomycetes. Nat. Prod. Rep. 2014, 31, 381–399. [Google Scholar] [CrossRef]
- Khan, S.T.; Komaki, H.; Motohashi, K.; Kozone, I.; Mukai, A.; Takagi, M.; Shin-ya, K. Streptomyces associated with a marine sponge Haliclona sp.; biosynthetic genes for secondary metabolites and products. Environ. Microbiol. 2011, 13, 391–403. [Google Scholar] [CrossRef]
- Balasubramanian, S.; Othman, E.M.; Kampik, D.; Stopper, H.; Hentschel, U.; Ziebuhr, W.; Oelschlaeger, T.A.; Abdelmohsen, U.R. Marine sponge-derived Streptomyces sp. SBT343 extract inhibits staphylococcal biofilm formation. Front. Microbiol. 2017, 8, 236. [Google Scholar] [CrossRef] [Green Version]
- Abdelmohsen, U.R.; Zhang, G.; Philippe, A.; Schmitz, W.; Pimentel-Elardo, S.M.; Hertlein-Amslinger, B.; Hentschel, U.; Bringmann, G. Cyclodysidins A–D, cyclic lipopeptides from the marine sponge-derived Streptomyces strain RV15. Tetrahedron Lett. 2012, 53, 23–29. [Google Scholar] [CrossRef]
- Lin, S.-C.; Lehman, C.W.; Stewart, A.K.; Panny, L.; Bracci, N.; Wright, J.L.; Paige, M.; Strangman, W.K.; Kehn-Hall, K. Homoseongomycin, a compound isolated from marine actinomycete bacteria K3-1, is a potent inhibitor of encephalitic alphaviruses. Antivir. Res. 2021, 191, 105087. [Google Scholar] [CrossRef]
- Mitova, M.; Popov, S.; De Rosa, S. Cyclic peptides from a Ruegeria strain of bacteria associated with the sponge Suberites domuncula. J. Nat. Prod. 2004, 67, 1178–1181. [Google Scholar] [CrossRef]
- Nagai, K.; Kamigiri, K.; Arao, N.; Suzumura, K.-I.; Kawano, Y.; Yamaoka, M.; Zhang, H.; Watanabe, M.; Suzuki, K. YM-266183 and ym-266184, novel thiopeptide antibiotics produced by Bacillus cereus isolated from a marine sponge i. taxonomy, fermentation, isolation, physico-chemical properties and biological properties. J. Antibiot. 2003, 56, 123–128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martín, J.; Sousa, D.S.; Crespo, G.; Palomo, S.; González, I.; Tormo, J.R.; De la Cruz, M.; Anderson, M.; Hill, R.T.; Vicente, F. Kocurin, the true structure of PM181104, an anti-methicillin-resistant Staphylococcus aureus (MRSA) thiazolyl peptide from the marine-derived bacterium Kocuria palustris. Mar. Drugs 2013, 11, 387–398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adnani, N.; Chevrette, M.G.; Adibhatla, S.N.; Zhang, F.; Yu, Q.; Braun, D.R.; Nelson, J.; Simpkins, S.W.; McDonald, B.R.; Myers, C.L. Coculture of marine invertebrate-associated bacteria and interdisciplinary technologies enable biosynthesis and discovery of a new antibiotic, keyicin. ACS Chem. Biol. 2017, 12, 3093–3102. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Li, Y.; Banakar, S.P.; Liu, L.; Shao, C.; Li, Z.; Wang, C. New metabolites from the co-culture of marine-derived actinomycete Streptomyces rochei MB037 and fungus Rhinocladiella similis 35. Front. Microbiol. 2019, 10, 915. [Google Scholar] [CrossRef] [Green Version]
- Dashti, Y.; Grkovic, T.; Abdelmohsen, U.R.; Hentschel, U.; Quinn, R.J. Production of induced secondary metabolites by a co-culture of sponge-associated actinomycetes, Actinokineospora sp. EG49 and Nocardiopsis sp. RV163. Mar. Drugs 2014, 12, 3046–3059. [Google Scholar] [CrossRef]
- Wu, Q.; Sun, J.; Chen, J.; Zhang, H.; Guo, Y.-W.; Wang, H. Terpenoids from marine soft coral of the genus Lemnalia: Chemistry and biological activities. Mar. Drugs 2018, 16, 320. [Google Scholar] [CrossRef] [Green Version]
- Webster, N.S.; Reusch, T.B. Microbial contributions to the persistence of coral reefs. ISME J. 2017, 11, 2167–2174. [Google Scholar] [CrossRef]
- Rosenberg, E.; Koren, O.; Reshef, L.; Efrony, R.; Zilber-Rosenberg, I. The role of microorganisms in coral health, disease and evolution. Nat. Rev. Microbiol. 2007, 5, 355–362. [Google Scholar] [CrossRef]
- Alagely, A.; Krediet, C.J.; Ritchie, K.B.; Teplitski, M. Signaling-mediated cross-talk modulates swarming and biofilm formation in a coral pathogen Serratia marcescens. ISME J. 2011, 5, 1609–1620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ritchie, K.B. Regulation of microbial populations by coral surface mucus and mucus-associated bacteria. Mar. Ecol. Prog. Ser. 2006, 322, 1–14. [Google Scholar] [CrossRef]
- Shnit-Orland, M.; Kushmaro, A. Coral mucus-associated bacteria: A possible first line of defense. FEMS Microbiol. Ecol. 2009, 67, 371–380. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, V.; Martín, J.; Sarmiento-Vizcaíno, A.; De la Cruz, M.; García, L.A.; Blanco, G.; Reyes, F. Anthracimycin B, a potent antibiotic against Gram-positive bacteria isolated from cultures of the deep-sea actinomycete Streptomyces cyaneofuscatus M-169. Mar. Drugs 2018, 16, 406. [Google Scholar] [CrossRef] [Green Version]
- Fu, P.; Kong, F.; Wang, Y.; Wang, Y.; Liu, P.; Zuo, G.; Zhu, W. Antibiotic metabolites from the coral-associated Actinomycete Streptomyces sp. OUCMDZ-1703. Chin. J. Chem. 2013, 31, 100–104. [Google Scholar] [CrossRef]
- Sarmiento-Vizcaíno, A.; González, V.; Braña, A.F.; Palacios, J.J.; Otero, L.; Fernández, J.; Molina, A.; Kulik, A.; Vázquez, F.; Acuña, J.L. Pharmacological potential of phylogenetically diverse Actinobacteria isolated from deep-sea coral ecosystems of the submarine Avilés Canyon in the Cantabrian Sea. Microb. Ecol. 2017, 73, 338–352. [Google Scholar] [CrossRef] [PubMed]
- Romero, F.; Espliego, F.; Baz, J.P.; De Quesada, T.G.; Gravalos, D.; De La Calle, F.; Fernandez-puentes, J.L. Thiocoraline, a new depsipeptide with antitumor activity produced by a marine Micromonospora I. Taxonomy, fermentation, isolation, and biological activities. J. Antibiot. 1997, 50, 734–737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benkendorff, K. Molluscan biological and chemical diversity: Secondary metabolites and medicinal resources produced by marine molluscs. Biol. Rev. 2010, 85, 757–775. [Google Scholar] [CrossRef]
- Wiese, J.; Abdelmohsen, U.R.; Motiei, A.; Humeida, U.H.; Imhoff, J.F. Bacicyclin, a new antibacterial cyclic hexapeptide from Bacillus sp. strain BC028 isolated from Mytilus edulis. Bioorg. Med. Chem. Lett. 2018, 28, 558–561. [Google Scholar] [CrossRef] [PubMed]
- Ji, X.; Dong, Y.; Ling, C.; Zhou, Z.; Li, Q.; Ju, J. Elucidation of the tailoring steps in Julichrome biosynthesis by marine gastropod mollusk-associated Streptomyces sampsonii SCSIO 054. Org. Lett. 2020, 22, 6927–6931. [Google Scholar] [CrossRef] [PubMed]
- Suria, A.M.; Tan, K.C.; Kerwin, A.H.; Gitzel, L.; Abini-Agbomson, L.; Bertenshaw, J.M.; Sewell, J.; Nyholm, S.V.; Balunas, M.J. Hawaiian bobtail squid symbionts inhibit marine bacteria via production of specialized metabolites, including new bromoalterochromides BAC-D/D′. Msphere 2020, 5, e00166-20. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez Jimenez, A.; Dechamps, E.; Giaux, A.; Goetghebuer, L.; Bauwens, M.; Willenz, P.; Flahaut, S.; Laport, M.S.; George, I.F. The sponges Hymeniacidon perlevis and Halichondria panicea are reservoirs of antibiotic-producing bacteria against multi-drug resistant Staphylococcus aureus. J. Appl. Microbiol. 2021, 131, 706–718. [Google Scholar] [CrossRef]
- Balansa, W.; Liu, Y.; Sharma, A.; Mihajlovic, S.; Hartwig, C.; Leis, B.; Rieuwpassa, F.J.; Ijong, F.G.; Wägele, H.; König, G.M. Selection of sponge-associated bacteria with high potential for the production of antibacterial compounds. Sci. Rep. 2020, 10, 19614. [Google Scholar]
- Bibi, F.; Yasir, M.; Al-Sofyani, A.; Naseer, M.I.; Azhar, E.I. Antimicrobial activity of bacteria from marine sponge Suberea mollis and bioactive metabolites of Vibrio sp. EA348. Saudi J. Biol. Sci. 2020, 27, 1139–1147. [Google Scholar] [CrossRef]
- Freitas-Silva, J.; Silva-Oliveira, T.; Muricy, G.; Laport, M.S. Bacillus strains associated to homoscleromorpha sponges are highly active against multidrug resistant bacteria. Curr. Microbiol. 2020, 77, 807–815. [Google Scholar] [CrossRef]
- Rajasabapathy, R.; Ghadi, S.C.; Manikandan, B.; Mohandass, C.; Surendran, A.; Dastager, S.G.; Meena, R.M.; James, R.A. Antimicrobial profiling of coral reef and sponge associated bacteria from southeast coast of India. Microb. Pathog. 2020, 141, 103972. [Google Scholar] [CrossRef]
- Braña, A.F.; Sarmiento-Vizcaíno, A.; Osset, M.; Pérez-Victoria, I.; Martín, J.; De Pedro, N.; De la Cruz, M.; Díaz, C.; Vicente, F.; Reyes, F. Lobophorin K, a new natural product with cytotoxic activity produced by Streptomyces sp. M-207 associated with the deep-sea coral Lophelia pertusa. Mar. Drugs 2017, 15, 144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shnit-Orland, M.; Sivan, A.; Kushmaro, A. Antibacterial activity of Pseudoalteromonas in the coral holobiont. Microb. Ecol. 2012, 64, 851–859. [Google Scholar] [CrossRef]
- Zhang, X.; Sun, Y.; Bao, J.; He, F.; Xu, X.; Qi, S. Phylogenetic survey and antimicrobial activity of culturable microorganisms associated with the South China Sea black coral Antipathes dichotoma. FEMS Microbiol. Lett. 2012, 336, 122–130. [Google Scholar] [CrossRef] [Green Version]
- Desriac, F.; El Harras, A.; Simon, M.; Bondon, A.; Brillet, B.; Le Chevalier, P.; Pugnière, M.; Got, P.; Destoumieux-Garzon, D.; Fleury, Y. Alterins produced by oyster-associated Pseudoalteromonas are antibacterial cyclolipopeptides with LPS-binding activity. Mar. Drugs 2020, 18, 630. [Google Scholar] [CrossRef]
- Dong, Y.; Ding, W.; Sun, C.; Ji, X.; Ling, C.; Zhou, Z.; Chen, Z.; Chen, X.; Ju, J. Julichrome monomers from marine gastropod mollusk-associated Streptomyces and stereochemical revision of Julichromes Q3⋅5 and Q3⋅3. Chem. Biodivers. 2020, 17, e2000057. [Google Scholar] [PubMed]
- Zhou, Z.; Wu, Q.; Xie, Q.; Ling, C.; Zhang, H.; Sun, C.; Ju, J. New borrelidins from Onchidium sp. associated Streptomyces olivaceus SCSIO LO13. Chem. Biodivers. 2020, 17, e1900560. [Google Scholar] [CrossRef] [PubMed]
- Lacerna, N.M.; Miller, B.W.; Lim, A.L.; Tun, J.O.; Robes, J.M.D.; Cleofas, M.J.B.; Lin, Z.; Salvador-Reyes, L.A.; Haygood, M.G.; Schmidt, E.W. Mindapyrroles A–C, pyoluteorin analogues from a shipworm-associated bacterium. J. Nat. Prod. 2019, 82, 1024–1028. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Koch, M.; Pond, C.D.; Mabeza, G.; Seronay, R.A.; Concepcion, G.P.; Barrows, L.R.; Olivera, B.M.; Schmidt, E.W. Structure and activity of lobophorins from a turrid mollusk-associated Streptomyces sp. J. Antibiot. 2014, 67, 121–126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalinovskaya, N.I.; Kalinovsky, A.I.; Romanenko, L.A.; Dmitrenok, P.S.; Kuznetsova, T.A. New angucyclines and antimicrobial diketopiperazines from the marine mollusk-derived actinomycete Saccharothrix espanaensis An 113. Nat. Prod. Commun. 2010, 5, 1934578X1000500420. [Google Scholar] [CrossRef] [Green Version]
- Romanenko, L.A.; Uchino, M.; Kalinovskaya, N.I.; Mikhailov, V.V. Isolation, phylogenetic analysis and screening of marine mollusc-associated bacteria for antimicrobial, hemolytic and surface activities. Microbiol. Res. 2008, 163, 633–644. [Google Scholar] [CrossRef]
- Fdhila, F.; Vázquez, V.; Sánchez, J.L.; Riguera, R. dd-Diketopiperazines: Antibiotics active against Vibrio a nguillarum isolated from marine bacteria associated with cultures of Pecten maximus. J. Nat. Prod. 2003, 66, 1299–1301. [Google Scholar] [CrossRef] [PubMed]
- Norziah, M.H.; Ching, C.Y. Nutritional composition of edible seaweed Gracilaria changgi. Food Chem. 2000, 68, 69–76. [Google Scholar] [CrossRef]
- Aryee, A.N.; Agyei, D.; Akanbi, T.O. Recovery and utilization of seaweed pigments in food processing. Curr. Opin. Food Sci. 2018, 19, 113–119. [Google Scholar] [CrossRef]
- Singh, R.P.; Reddy, C. Seaweed–microbial interactions: Key functions of seaweed-associated bacteria. FEMS Microbiol. Ecol. 2014, 88, 213–230. [Google Scholar] [CrossRef]
- Menaa, F.; Wijesinghe, P.; Thiripuranathar, G.; Uzair, B.; Iqbal, H.; Khan, B.A.; Menaa, B. Ecological and industrial implications of dynamic seaweed-associated microbiota interactions. Mar. Drugs 2020, 18, 641. [Google Scholar] [CrossRef] [PubMed]
- Lachnit, T.; Meske, D.; Wahl, M.; Harder, T.; Schmitz, R. Epibacterial community patterns on marine macroalgae are host-specific but temporally variable. Environ. Microbiol. 2011, 13, 655–665. [Google Scholar] [CrossRef] [PubMed]
- Burke, C.; Thomas, T.; Lewis, M.; Steinberg, P.; Kjelleberg, S. Composition, uniqueness and variability of the epiphytic bacterial community of the green alga Ulva australis. ISME J. 2011, 5, 590–600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hollants, J.; Leliaert, F.; Verbruggen, H.; Willems, A.; De Clerck, O. Permanent residents or temporary lodgers: Characterizing intracellular bacterial communities in the siphonous green alga Bryopsis. Proc. R. Soc. B Biol. Sci. 2013, 280, 20122659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gozari, M.; Alborz, M.; El-Seedi, H.R.; Jassbi, A.R. Chemistry, biosynthesis and biological activity of terpenoids and meroterpenoids in bacteria and fungi isolated from different marine habitats. Eur. J. Med. Chem. 2020, 210, 112957. [Google Scholar] [CrossRef] [PubMed]
- Rungprom, W.; Siwu, E.R.; Lambert, L.K.; Dechsakulwatana, C.; Barden, M.C.; Kokpol, U.; Blanchfield, J.T.; Kita, M.; Garson, M.J. Cyclic tetrapeptides from marine bacteria associated with the seaweed Diginea sp. and the sponge Halisarca ectofibrosa. Tetrahedron 2008, 64, 3147–3152. [Google Scholar] [CrossRef]
- Franks, A.; Haywood, P.; Holmström, C.; Egan, S.; Kjelleberg, S.; Kumar, N. Isolation and structure elucidation of a novel yellow pigment from the marine bacterium Pseudoalteromonas tunicata. Molecules 2005, 10, 1286–1291. [Google Scholar] [CrossRef] [Green Version]
- Franks, A.; Egan, S.; Holmström, C.; James, S.; Lappin-Scott, H.; Kjelleberg, S. Inhibition of fungal colonization by Pseudoalteromonas tunicata provides a competitive advantage during surface colonization. Appl. Environ. Microbiol. 2006, 72, 6079–6087. [Google Scholar] [CrossRef] [Green Version]
- Gandhi, N.; Nazareth, J.; Divekar, P.; Kohl, H.; De Souza, N. Magnesidin, a novel magnesium-containing antibiotic. J. Antibiot. 1973, 26, 797–798. [Google Scholar] [CrossRef] [Green Version]
- Isnansetyo, A.; Horikawa, M.; Kamei, Y. In vitro anti-methicillin-resistant Staphylococcus aureus activity of 2, 4-diacetylphloroglucinol produced by Pseudomonas sp. AMSN isolated from a marine alga. J. Antimicrob. Chemother. 2001, 47, 724–725. [Google Scholar] [CrossRef] [Green Version]
- Ravisankar, A.; Gnanambal, M.E.K.; Sundaram, L.R. A newly isolated Pseudomonas sp., epibiotic on the seaweed, Padina tetrastromatica, off Southeastern Coast of India, reveals antibacterial action. Appl. Biochem. Biotechnol. 2013, 171, 1968–1985. [Google Scholar] [CrossRef]
- Imamura, N.; Nishijima, M.; Takadera, T.; Adachi, K.; SAKAI, M.; Sano, H. New anticancer antibiotics pelagiomicins, produced by a new marine bacterium Pelagiobacter variabilis. J. Antibiot. 1997, 50, 8–12. [Google Scholar] [CrossRef] [Green Version]
- Short, F.T.; Polidoro, B.; Livingstone, S.R.; Carpenter, K.E.; Bandeira, S.; Bujang, J.S.; Calumpong, H.P.; Carruthers, T.J.; Coles, R.G.; Dennison, W.C. Extinction risk assessment of the world’s seagrass species. Biol. Conserv. 2011, 144, 1961–1971. [Google Scholar] [CrossRef]
- Bertelli, C.M.; Unsworth, R.K. Protecting the hand that feeds us: Seagrass (Zostera marina) serves as commercial juvenile fish habitat. Mar. Pollut. Bull. 2014, 83, 425–429. [Google Scholar] [CrossRef]
- Custódio, L.; Laukaityte, S.; Engelen, A.H.; Rodrigues, M.J.; Pereira, H.; Vizetto-Duarte, C.; Barreira, L.; Rodríguez, H.; Alberício, F.; Varela, J. A comparative evaluation of biological activities and bioactive compounds of the seagrasses Zostera marina and Zostera noltei from southern Portugal. Nat. Prod. Res. 2016, 30, 724–728. [Google Scholar] [CrossRef] [Green Version]
- Rotter, A.; Barbier, M.; Bertoni, F.; Bones, A.M.; Cancela, M.L.; Carlsson, J.; Carvalho, M.F.; Cegłowska, M.; Chirivella-Martorell, J.; Conk Dalay, M. The essentials of marine biotechnology. Front. Mar. Sci. 2021, 8, 158. [Google Scholar] [CrossRef]
- Bibi, F.; Naseer, M.I.; Hassan, A.M.; Yasir, M.; Al-Ghamdi, A.A.K.; Azhar, E.I. Diversity and antagonistic potential of bacteria isolated from marine grass Halodule uninervis. 3 Biotech 2018, 8, 48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boontanom, P.; Chantarasiri, A. Diversity of culturable epiphytic bacteria isolated from seagrass (Halodule uninervis) in Thailand and their preliminary antibacterial activity. Biodiversitas J. Biol. Divers. 2020, 21, 7. [Google Scholar] [CrossRef]
- Thatoi, H.; Biswal, A. Mangroves of Orissa Coast: Floral diversity and conservation status. Spec. Habit. Threaten. Plants India ENVIS Wild Life Protect. Area 2008, 11, 201–207. [Google Scholar]
- Sarma, V.; Hyde, K.D. A review on frequently occurring fungi in mangroves. Fungal Divers. 2001, 8, 1–34. [Google Scholar]
- Hong, K.; Gao, A.-H.; Xie, Q.-Y.; Gao, H.G.; Zhuang, L.; Lin, H.-P.; Yu, H.-P.; Li, J.; Yao, X.-S.; Goodfellow, M. Actinomycetes for marine drug discovery isolated from mangrove soils and plants in China. Mar. Drugs 2009, 7, 24–44. [Google Scholar] [CrossRef] [PubMed]
- Sivakumar, K. Actinomycetes of an Indian Mangrove (Pitchavaram) Environment; an Inventory. Ph.D. Thesis, Annamalai University, Annamalai Nagar, India, 2001. [Google Scholar]
- Laksmanaperumalsamy, P.; Chandramohan, D.; Natarajan, R. Antibacterial and antifungal activity of streptomycetes from Porto Novo coastal environment. Mar. Biol. 1978, 11, 15–24. [Google Scholar]
- Eccleston, G.P.; Brooks, P.R.; Kurtböke, D.I. The occurrence of bioactive micromonosporae in aquatic habitats of the Sunshine Coast in Australia. Mar. Drugs 2008, 6, 243–261. [Google Scholar] [CrossRef]
- Huang, H.; Lv, J.; Hu, Y.; Fang, Z.; Zhang, K.; Bao, S. Micromonospora rifamycinica sp. nov., a novel actinomycete from mangrove sediment. Int. J. Syst. Evol. Microbiol. 2008, 58, 17–20. [Google Scholar] [CrossRef]
- Xie, X.-C.; Mei, W.-L.; Zhao, Y.-X.; Hong, K.; Dai, H.-F. A new degraded sesquiterpene from marine actinomycete Streptomyces sp. 0616208. Chin. Chem. Lett. 2006, 17, 1463–1465. [Google Scholar]
- Retnowati, W. Identification of Streptomyces sp-MWS1 producing antibacterial compounds. Indones. J. Trop. Infect. Dis. 2010, 1, 82–85. [Google Scholar] [CrossRef]
- Satheeja, S.V.; Jebakumar, S.R. Phylogenetic analysis and antimicrobial activities of Streptomyces isolates from mangrove sediment. J. Basic Microbiol. 2011, 51, 71–79. [Google Scholar] [CrossRef]
- Karthick, P.; Mohanraju, R. Antimicrobial compounds produced by Lysinibacillus odysseyi epiphytic bacteria associated with red algae. Braz. J. Microbiol. 2020, 51, 1683–1690. [Google Scholar] [CrossRef] [PubMed]
- Horta, A.; Alves, C.; Pinteus, S.; Lopes, C.; Fino, N.; Silva, J.; Ribeiro, J.; Rodrigues, D.; Francisco, J.; Rodrigues, A. Identification of Asparagopsis armata-associated bacteria and characterization of their bioactive potential. Microbiol. Open 2019, 8, e00824. [Google Scholar] [CrossRef] [PubMed]
- Karthick, P.; Mohanraju, R. Antimicrobial potential of epiphytic bacteria associated with seaweeds of Little Andaman, India. Front. Microbiol. 2018, 9, 611. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Braña, A.F.; Sarmiento-Vizcaíno, A.; Pérez-Victoria, I.; Martín, J.; Otero, L.; Palacios-Gutiérrez, J.J.; Fernández, J.; Mohamedi, Y.; Fontanil, T.; Salmón, M. Desertomycin G, a new antibiotic with activity against Mycobacterium tuberculosis and human breast tumor cell lines produced by Streptomyces althioticus MSM3, isolated from the Cantabrian Sea Intertidal macroalgae Ulva sp. Mar. Drugs 2019, 17, 114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chakraborty, K.; Thilakan, B.; Kizhakkekalam, V. Antibacterial aryl-crowned polyketide from Bacillus subtilis associated with seaweed Anthophycus longifolius. J. Appl. Microbiol. 2018, 124, 108–125. [Google Scholar] [CrossRef] [PubMed]
- Uzair, B.; Menaa, F.; Khan, B.A.; Mohammad, F.V.; Ahmad, V.U.; Djeribi, R.; Menaa, B. Isolation, purification, structural elucidation and antimicrobial activities of kocumarin, a novel antibiotic isolated from actinobacterium Kocuria marina CMG S2 associated with the brown seaweed Pelvetia canaliculata. Microbiol. Res. 2018, 206, 186–197. [Google Scholar] [CrossRef]
- Chakraborty, K.; Thilakan, B.; Raola, V.K. Previously undescribed antibacterial polyketides from heterotrophic Bacillus amyloliquefaciens associated with seaweed Padina gymnospora. Appl. Biochem. Biotechnol. 2018, 184, 716–732. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, K.; Thilakan, B.; Raola, V.K. Antimicrobial polyketide furanoterpenoids from seaweed-associated heterotrophic bacterium Bacillus subtilis MTCC 10403. Phytochemistry 2017, 142, 112–125. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, K.; Thilakan, B.; Chakraborty, R.D.; Raola, V.K.; Joy, M. O-heterocyclic derivatives with antibacterial properties from marine bacterium Bacillus subtilis associated with seaweed, Sargassum myriocystum. Appl. Microbiol. Biotechnol. 2017, 101, 569–583. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, K.; Thilakan, B.; Raola, V.K.; Joy, M. Antibacterial polyketides from Bacillus amyloliquefaciens associated with edible red seaweed Laurenciae papillosa. Food Chem. 2017, 218, 427–434. [Google Scholar] [CrossRef]
- Thilakan, B.; Chakraborty, K.; Chakraborty, R.D. Antimicrobial properties of cultivable bacteria associated with seaweeds in the Gulf of Mannar on the southeast coast of India. Can. J. Microbiol. 2016, 62, 668–681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ismail, A.; Ktari, L.; Ahmed, M.; Bolhuis, H.; Boudabbous, A.; Stal, L.J.; Cretoiu, M.S.; El Bour, M. Antimicrobial activities of bacteria associated with the brown alga Padina pavonica. Front. Microbiol. 2016, 7, 1072. [Google Scholar] [CrossRef]
- Chakraborty, K.; Thilakan, B.; Raola, V.K. Polyketide family of novel antibacterial 7-O-methyl-5′-hydroxy-3′-heptenoate–macrolactin from seaweed-associated Bacillus subtilis MTCC 10403. J. Agric. Food Chem. 2014, 62, 12194–12208. [Google Scholar] [CrossRef]
- Jamal, M.T.; Morris, P.C.; Hansen, R.; Jamieson, D.J.; Burgess, J.G.; Austin, B. Recovery and characterization of a 30.7-kDa protein from Bacillus licheniformis associated with inhibitory activity against methicillin-resistant Staphylococcus aureus, vancomycin-resistant Enterococci, and Listeria monocytogenes. Mar. Biotechnol. 2006, 8, 587–592. [Google Scholar] [CrossRef]
- Cristianawati, O.; Sibero, M.T.; Ayuningrum, D.; Nuryadi, H.; Syafitri, E.; Riniarsih, I.; Radjasa, O.K. Screening of antibacterial activity of seagrass-associated bacteria from the North Java Sea, Indonesia against multidrug-resistant bacteria. Aquac. Aquar. Conserv. Legislat. 2019, 12, 1054–1064. [Google Scholar]
- Ravikumar, S.; Thajuddin, N.; Suganthi, P.; Jacob Inbaneson, S.; Vinodkumar, T. Bioactive potential of seagrass bacteria against human bacterial pathogens. J. Environ. Biol. 2010, 31, 387. [Google Scholar]
- Bodhaguru, M.; Prakash, S.; Ramasubburayan, R.; Ahila, N.K.; Mariselvam, L.; Immanuel, G.; Palavesam, A.; Kannapiran, E. Screening, partial purification of antivibriosis metabolite sterol-glycosides from Rhodococcus sp. against aquaculture associated pathogens. Microb. Pathog. 2019, 134, 103597. [Google Scholar] [CrossRef]
- Bibi, F.; Ullah, I.; Alvi, S.; Bakhsh, S.; Yasir, M.; Al-Ghamdi, A.; Azhar, E. Isolation, diversity, and biotechnological potential of rhizo-and endophytic bacteria associated with mangrove plants from Saudi Arabia. Genet. Mol. Res. 2017, 16, gmr16029657. [Google Scholar] [CrossRef]
- Balakrishnan, B.; Ranishree, J.K.; Thadikamala, S.; Panchatcharam, P. Purification, characterization and production optimization of a vibriocin produced by mangrove associated Vibrio parahaemolyticus. Asian Pac. J. Trop. Biomed. 2014, 4, 253–261. [Google Scholar] [CrossRef] [Green Version]
- Bhadury, P.; Mohammad, B.T.; Wright, P.C. The current status of natural products from marine fungi and their potential as anti-infective agents. J. Ind. Microbiol. Biotechnol. 2006, 33, 325. [Google Scholar] [CrossRef] [PubMed]
- Jensen, P.R. Natural products and the gene cluster revolution. Trends Microbiol. 2016, 24, 968–977. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiang, Y.-M.; Chang, S.-L.; Oakley, B.R.; Wang, C.C. Recent advances in awakening silent biosynthetic gene clusters and linking orphan clusters to natural products in microorganisms. Curr. Opin. Chem. Biol. 2011, 15, 137–143. [Google Scholar] [CrossRef] [Green Version]
- Konstantinou, D.; Mavrogonatou, E.; Zervou, S.-K.; Giannogonas, P.; Gkelis, S. Bioprospecting sponge-associated marine Cyanobacteria to produce bioactive compounds. Toxins 2020, 12, 73. [Google Scholar] [CrossRef] [PubMed]
- Graça, A.P.; Calisto, R.; Lage, O.M. Planctomycetes as novel source of bioactive molecules. Front. Microbiol. 2016, 7, 1241. [Google Scholar] [CrossRef] [PubMed]
- Wade, W. Unculturable bacteria—The uncharacterized organisms that cause oral infections. J. R. Soc. Med. 2002, 95, 81–83. [Google Scholar] [PubMed]
- Vartoukian, S.R.; Palmer, R.M.; Wade, W.G. Strategies for culture of ‘unculturable’bacteria. FEMS Microbiol. Lett. 2010, 309, 1–7. [Google Scholar] [PubMed] [Green Version]
- Zeaiter, Z.; Mapelli, F.; Crotti, E.; Borin, S. Methods for the genetic manipulation of marine bacteria. Electron. J. Biotechnol. 2018, 33, 17–28. [Google Scholar] [CrossRef]
- Trindade, M.; Van Zyl, L.J.; Navarro-Fernández, J.; Abd Elrazak, A. Targeted metagenomics as a tool to tap into marine natural product diversity for the discovery and production of drug candidates. Front. Microbiol. 2015, 6, 890. [Google Scholar] [CrossRef] [PubMed]
- Mahapatra, G.P.; Raman, S.; Nayak, S.; Gouda, S.; Das, G.; Patra, J.K. Metagenomics approaches in discovery and development of new bioactive compounds from marine actinomycetes. Curr. Microbiol. 2019, 77, 645–656. [Google Scholar] [CrossRef]
- Olano, C.; García, I.; González, A.; Rodriguez, M.; Rozas, D.; Rubio, J.; Sánchez-Hidalgo, M.; Braña, A.F.; Méndez, C.; Salas, J.A. Activation and identification of five clusters for secondary metabolites in Streptomyces albus J 1074. Microb. Biotechnol. 2014, 7, 242–256. [Google Scholar] [CrossRef] [PubMed] [Green Version]















| S. No | Marine Source | Marine Bacteria | Secondary Metabolite(s) | Antimicrobial Activity | Ref |
|---|---|---|---|---|---|
| 1 | Sediment sample | Nonomuraea sp. MM565M-173N2 | Sealutomicin A | Inhibited the growth of carbapenem-resistant Enterobacteriaceae | [155] |
| 2 | Sediment sample | Streptomyces sp. | - | Exhibited antifungal activity against C. albicans | [156] |
| 3 | Sediment sample | Actinomycete AMA50 | Tetradecanoic acid, pentadecanoic acid, and n-hexadecanoic acid | Exhibited antifungal activity against Talaromyces marneffei | [157] |
| 4 | Sediment sample | Streptomyces sp. SY1965 | Streptothiazolidine A | Exhibited antifungal activity against C. albicans | [158] |
| 5 | Sediment sample | B. velezensis SH-B74 | Anteiso-C15 Ile2,7 surfactin, 1 | Inhibited the appressoria formation of rice blast fungal pathogen Magnaporthe oryzae | [159] |
| 6 | Sediment sample | Pseudomonas sp. | Chitinase | Showed antifungal activity against Verticillium dahlia CICC 2534 and Fusarium oxysporum f. sp. cucumerinum CICC 2532 | [160] |
| 7 | Sediment sample | Brevibacillus antibioticus sp. TGS2-1T | Different fatty acids and lipid compounds | Exhibited both antibacterial and antifungal activities | [161] |
| 8 | Sediment sample | Nocardiopsis sp. SCA21 | 4-bromophenol and bis (2-ethylhexyl) phthalate | Showed antibacterial activity against several Gram-positive and Gram-negative bacteria | [162] |
| 9 | Sediment sample | Salinispora arenicola | 3-hydroxy-N-methyl-2-oxindole derivatives | Inhibited the growth of E. faecalis | [163] |
| 10 | Estuary soil sample | S. felleus | Polyketide compounds | Exhibited antibacterial activity against Enterococcus sp. | [164] |
| 11 | Sediment sample | S. arenicola | Salinaphthoquinones | Exhibited moderate antibacterial activity against E. faecalis and S. aureus | [165] |
| 12 | Sediment sample | Streptomyces sp. | Meroterpenoids | Showed intense antibacterial activity against B. subtilis and S. aureus | [166] |
| 13 | Sediment sample | Streptomyces sp. RKND004 | Terrosamycins A and B | Exhibited strong antibacterial activity against Gram-positive bacteria | [167] |
| 14 | Sediment sample | Streptomyces strains | - | Exhibited antifungal activity against C. albicans and A. niger | [168] |
| 15 | Sediment sample | Streptomyces sp. 4054 | Mycenolide A | Inhibited the growth of B. subtilis | [169] |
| 16 | Sediment sample | S. xinghaiensis SCSIO S15077 | Tunicamycin derivatives | Showed both antibacterial and antifungal activities | [170] |
| 17 | Sediment sample | Bacillus and Virgibacillus strains | - | Exhibited antibacterial activity against S. aureus and V. parahaemolyticus | [171] |
| 18 | Sediment sample | Actinobacteria SJP4 | [1,2,4]triazol-1-ylethanone | Exhibited antibacterial activity against both Gram-positive and Gram-negative bacteria | [172] |
| 19 | Sediment sample | Streptomyces sp. Bacillus sp. and Micrococcus sp. | - | Showed both antibacterial and antifungal activities | [173] |
| 20 | Sediment sample | S. mutabilis sp. MII | N-acetylborrelidin B | Exhibited both antibacterial and antifungal activities | [174] |
| 21 | Mangrove sediment sample | Streptomyces | - | Exhibited antibacterial activity against both Gram-positive and Gram-negative bacteria | [175] |
| 22 | Mangrove sediment sample | Actinobacteria | Extracellular enzymes | Exhibited both antibacterial and antifungal activities | [176] |
| 23 | Sediment sample | Aneurinibacillus sp. YR247 | Peptidic compounds | Showed antifungal activity against A. brasiliensis NBRC945 | [177] |
| 24 | Sediment sample | Rheinheimera japonica KMM 9513 T | Diketopiperazines | Inhibited the growth of B. subtilis, S. aureus and E. faecium | [178] |
| 25 | Sediment sample | T. flavus SP5 | Different bioactive compounds | Inhibited growth of both bacterial and fungal pathogens | [179] |
| 26 | Mangrove sediment sample | S. parvulus DOSMB-D105 | Different bioactive compounds | Exhibited both antibacterial and antifungal activities | [180] |
| 27 | Sediment sample | R. japonica sp. | Isoprenoid quinones | Exhibited strong antagonistic activity against both Gram-positive and Gram-negative bacteria | [181] |
| 28 | Sediment sample | Streptomyces sp. CMB-M0244 | Mollemycin A | Exhibited antibacterial activity against both Gram-positive and Gram-negative bacteria | [182] |
| 29 | Sediment sample | B. sonorensis MT93 | Sonorensin | Exhibited broad spectrum of antibacterial activity against different bacterial pathogens | [183] |
| 30 | Sediment sample | Streptosporangium sp. DSM 45942 | Iodinin | Exhibited both antibacterial and antifungal activities | [184] |
| 31 | Sediment sample | Streptomyces sp. 12A35 | Lobophorin I | Exhibited antibacterial activity against S. aureus and B. subtilis | [185] |
| 32 | Sediment sample | Actinoalloteichus sp. NPS702 | Neomaclafungins | Showed strong antifungal activity against Trichophyton mentagrophytes | [186] |
| 33 | Sediment sample | Pseudonocardia sp. SCSIO 01299 | Pseudonocardians A–C | Exhibited antibacterial activity against both Gram-positive and Gram-negative bacteria | [187] |
| 34 | Sediment sample | Streptomyces sp. NTK 937 | Caboxamycin | Exhibited antibacterial activity against Gram-positive bacterial pathogens | [188] |
| 35 | Sediment sample | B. laterosporus Lh-1 | - | Exhibited antibacterial activity against both Gram-positive and Gram-negative bacteria | [189] |
| 36 | Sediment sample | Streptomyces sp. BD21-2 | Bonactin | Exhibited antibacterial activity against both Gram-positive and Gram-negative bacteria | [190] |
| 37 | Sediment sample | S. koyangensis SCSIO 5802 | Neoabyssomicins | Exhibited antiviral activity against HSV and vesicular stomatitis virus | [191] |
| 38 | Sediment sample | Streptomyces. sp. #HK18 | Xiamycin C | Exhibited strong antiviral activity against porcine epidemic diarrhea virus | [192] |
| 39 | Water sample | P. putida | 9, 10-dihydrophenanthrene-2-carboxylic acid | Revealed strong antifungal activity against C. albicans | [193] |
| 40 | Water sample | Bacillus, Arthrobacter, and Brevundimonas | - | Exhibited antibacterial activity against both Gram-positive and Gram-negative bacteria | [194] |
| 41 | Water sample | Pseudoalteromonas haloplanktis TAC125 | Methylamine | Inhibited the growth of Burkholderia cepacia complex | [195] |
| 42 | Estuarine water sample | Different strains of heterotrophic bacteria | - | Inhibited the growth of S. aureus and E. coli | [196] |
| S. No | Marine Fauna | Associated Marine Bacteria | Secondary Metabolite(s) | Antimicrobial Activity | Ref |
|---|---|---|---|---|---|
| 1 | Sponges (H. panacea and Hymeniacidon perlevis) | Maribacter, Aquimarina, Vagococcus, and Denitrobaculum bacterial isolates | - | Exhibited antibacterial activity against S. aureus | [228] |
| 2 | Sponges (Agelas nakamurai and Aaptos suberitoides) | Different Bacillus sp. | Macrolactin A and C14-surfactin | Inhibited the growth of E. coli and M. luteus | [229] |
| 3 | Sponge (Suberea mollis) | Vibrio sp. EA348 | Fosfomycin and amifloxacin | Exhibited antibacterial activity against both Gram-positive and Gram-negative bacteria | [230] |
| 4 | Sponges (Demospongiae and Homoscleromorpha) | Genera Vibrio and Bacillus | - | Inhibited the growth of different multidrug-resistant bacterial pathogens | [231] |
| 5 | Sponge (Arenosclera brasiliensis) | Diversity of heterotrophic bacteria | - | Exhibited antibacterial activity against B. subtilis | [231] |
| 6 | Sponge (Orina sagittaria) | Actinomycete strain F-04 | Volatile organic compounds | Inhibited growth of S. aureus | [232] |
| 7 | Coral (Lophelia pertusa) | Streptomyces sp. M-207 | Lobophorin K | Exhibited antibacterial activity against S. aureus | [233] |
| 8 | Coral (Platygyra sp.) | Pseudoalteromonas sp. | Heat tolerant cell-free culture supernatant | Inhibited the growth of B. cereus and S. aureus | [234] |
| 9 | Coral (Antipathes dichotoma) | Three different bacterial phyla (Actinobacteria, Alphaproteobacteria, and Firmicutes) | - | Exhibited both antibacterial and antifungal activities | [235] |
| 10 | Mollusk (Oysters—Crassostrea gigas) | Pseudoalteromonas hCg-6 and hCg-42 | Cyclolipopeptides | Inhibited the growth of Gram-negative human bacterial pathogens | [236] |
| 11 | Mollusk (Batillaria zonalis) | S. sampsonii SCSIO 054 | Julichrome Monomers | Inhibited the growth of S. simulans and S. aureus | [237] |
| 12 | Mollusk (Onchidium sp.) | S. olivaceus SCSIO LO13 | Borrelidins | Exhibited antibacterial activity against both Gram-positive and Gram-negative bacterial pathogens | [238] |
| 13 | Mollusk (Kuphus polythalamius) | P. aeruginosa 1682U.R.0a.27 | Mindapyrroles A–C | Showed antibacterial activity against both Gram-positive and Gram-negative bacteria | [239] |
| 14 | Mollusk (M. edulis) | Bacillus sp. BC028 | Bacicyclin | Inhibited the growth of clinical pathogens S. aureus and E. faecalis | [225] |
| 15 | Mollusk (Lienardia totopotens) | Streptomyces sp. | Lobophorins | Inhibited the growth of M. tuberculosis and B. cepacia | [240] |
| 16 | Mollusk (Anadara broughtoni) | Saccharothrix espanaensis An 113 | Saccharothrixmicines A and B | Exhibited antifungal activity against C. albicans | [241] |
| 17 | Mollusk (A. broughtoni) | B. pumilus An 112 | Cyclic depsipeptides | Exhibited broad spectrum antibacterial activity | [242] |
| 18 | Mollusk (Pecten maximus) | Bacterial strains CF-20 and C-148 | dd-diketopiperazines | Exhibited antibacterial activity against V. anguillarum | [243] |
| S. No | Marine Flora | Associated Marine Bacteria | Secondary Metabolite(s) | Antimicrobial Activity | Ref |
|---|---|---|---|---|---|
| 1 | Seaweed (Gracilaria canaliculata) | Lysinibacillus odysseyi KC149512 | Lupenol, diazene, and furan | Inhibited the growth of Gram-negative bacterial pathogens | [275] |
| 2 | Seaweed (Asparagopsis armata) | Shewanella sp. ASP 26 | - | Showed antibacterial activity against S. aureus and B. subtilis | [276] |
| 3 | Seaweeds (U. lactuca, G. corticata, and Mastophora rosea) | Phyla of Proteobacteria and Firmicutes | 2-Pyrrolidinone, Phenol, 2, 4-bis (1, 1-dimethylethyl) and Furan derivatives | Exhibited antibacterial activity against clinical pathogens | [277] |
| 4 | Seaweed (Ulva sp.) | S. althioticus MSM3 | Desertomycin G | Showed antibacterial activity against a broad range of Gram-positive bacterial pathogens | [278] |
| 5 | Seaweed (Anthophycus longifolius) | B. subtilis MTCC 10403 | Aryl-crowned polyketides | Exhibited antibacterial activity against different Gram-negative bacterial pathogens | [279] |
| 6 | Seaweed (Pelvetia canaliculata) | K. marina CMG S2 | Kocumarin | Inhibited the growth of both bacterial and fungal pathogens | [280] |
| 7 | Seaweed (P. gymnospora) | B. amyloliquefaciens | Polyketides | Exhibited antibacterial activity against V. vulnificus and V. parahaemolyticus | [281] |
| 8 | Seaweed (A. longifolius) | B. subtilis MTCC 10403 | Polyketide furanoterpenoids | Exhibited antibacterial activity against perceptive food pathogens | [282] |
| 9 | Seaweed (Sargassum myriocystum) | B. subtilis MTCC 10407 | O-heterocyclic polyketide derivatives | Showed potent antibacterial activity against V. parahaemolyticus, Aeromonas hydrophila, and V. vulnificus | [283] |
| 10 | Seaweed (Laurenciae papillosa) | B. amyloliquefaciens | Polyketides | Inhibited the growth of food-borne pathogens | [284] |
| 11 | Seaweed (Rhodophyceae and Phaeophyceae) | Phyla of Firmicutes and Proteobacteria | Polyketides | Prevented the growth of fouling bacteria | [285] |
| 12 | Seaweed (P. pavonica) | B. pumilus P8 | - | Exhibited both antibacterial and antifungal activities | [286] |
| 13 | Seaweed (A. longifolius) | B. subtilis MTCC 10403 | 7-O-methyl-5′-hydroxy-3′-heptenoate-macrolactin | Exhibited antibacterial activity against human opportunistic clinical pathogens | [287] |
| 14 | Seaweed (Fucus serratus) | B. licheniformis | YbdN protein | Inhibited the growth of S. aureus and Listeria monocytogenes | [288] |
| 15 | Seagrasses (Cymodocea sp., Enhalus acoroides, Syringodium sp., and Thalassia hemprichii) | B. flexus EED 15 and S. lienomycini EED 16 | - | Inhibited the growth of E. coli and S. aureus | [289] |
| 16 | Seagrasses (Cymodocea serrulata and Syringodium isoetifolium) | Different endo and epiphytic bacteria | - | Inhibited the growth of different human bacterial pathogens | [290] |
| 17 | Mangrove (Rhizophora mucronata) | Rhodococcus sp. | Sterol-glycosides | Exhibited antibacterial activity against aquatic bacterial pathogens | [291] |
| 18 | Mangroves (Seven different) | Belongs to the phylum Gammaproteobacteria | - | Showed antifungal activity against fungal pathogens | [292] |
| 19 | Mangrove (Avicennia marina) | V. parahaemolyticus | Vibriocin | Used in the management of controlling the vibrio infections | [293] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Srinivasan, R.; Kannappan, A.; Shi, C.; Lin, X. Marine Bacterial Secondary Metabolites: A Treasure House for Structurally Unique and Effective Antimicrobial Compounds. Mar. Drugs 2021, 19, 530. https://0-doi-org.brum.beds.ac.uk/10.3390/md19100530
Srinivasan R, Kannappan A, Shi C, Lin X. Marine Bacterial Secondary Metabolites: A Treasure House for Structurally Unique and Effective Antimicrobial Compounds. Marine Drugs. 2021; 19(10):530. https://0-doi-org.brum.beds.ac.uk/10.3390/md19100530
Chicago/Turabian StyleSrinivasan, Ramanathan, Arunachalam Kannappan, Chunlei Shi, and Xiangmin Lin. 2021. "Marine Bacterial Secondary Metabolites: A Treasure House for Structurally Unique and Effective Antimicrobial Compounds" Marine Drugs 19, no. 10: 530. https://0-doi-org.brum.beds.ac.uk/10.3390/md19100530