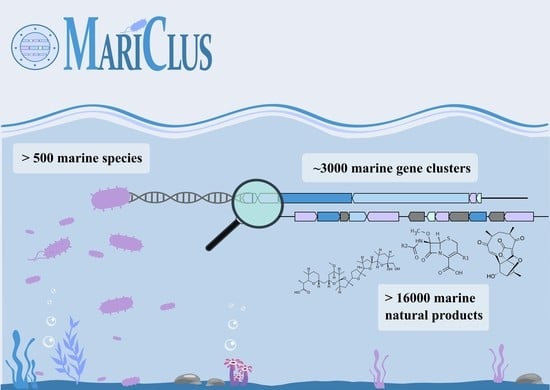
MariClus: Your One-Stop Platform for Information on Marine Natural Products, Their Gene Clusters and Producing Organisms
Abstract
:1. Introduction
2. Results
2.1. Data Collection
2.2. Online Portal
2.3. Meta-Analyses
2.3.1. Taxonomical Statistics
2.3.2. Cluster Types
2.3.3. Molecule Types
2.4. Case Studies
2.4.1. Microbiology
2.4.2. Gene Cluster Identification
2.4.3. Natural Product Discovery
3. Discussion
4. Materials and Methods
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Seyedsayamdost, M.R. Toward a Global Picture of Bacterial Secondary Metabolism. J. Ind. Microbiol. Biotechnol. 2019, 46, 301–311. [Google Scholar] [CrossRef] [PubMed]
- Arnison, P.G.; Bibb, M.J.; Bierbaum, G.; Bowers, A.A.; Bugni, T.S.; Bulaj, G.; Camarero, J.A.; Campopiano, D.J.; Challis, G.L.; Clardy, J.; et al. Ribosomally Synthesized and Post-Translationally Modified Peptide Natural Products: Overview and Recommendations for a Universal Nomenclature. Nat. Prod. Rep. 2013, 30, 108–160. [Google Scholar] [CrossRef] [PubMed]
- Medema, M.H.; Kottmann, R.; Yilmaz, P.; Cummings, M.; Biggins, J.B.; Blin, K.; de Bruijn, I.; Chooi, Y.H.; Claesen, J.; Coates, R.C.; et al. Minimum Information about a Biosynthetic Gene Cluster. Nat. Chem. Biol. 2015, 11, 625–631. [Google Scholar] [CrossRef]
- De Rop, A.-S.; Rombaut, J.; Willems, T.; De Graeve, M.; Vanhaecke, L.; Hulpiau, P.; De Maeseneire, S.L.; De Mol, M.L.; Soetaert, W.K. Novel Alkaloids from Marine Actinobacteria: Discovery and Characterization. Mar. Drugs 2022, 20, 6. [Google Scholar] [CrossRef]
- Pan, S.-Y.; Zhou, S.-F.; Gao, S.-H.; Yu, Z.-L.; Zhang, S.-F.; Tang, M.-K.; Sun, J.-N.; Ma, D.-L.; Han, Y.-F.; Fong, W.-F.; et al. New Perspectives on How to Discover Drugs from Herbal Medicines: CAM’s Outstanding Contribution to Modern Therapeutics. Evid. Based Complement. Altern. Med. 2013, 2013, e627375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiménez, C. Marine Natural Products in Medicinal Chemistry. ACS Med. Chem. Lett. 2018, 9, 959–961. [Google Scholar] [CrossRef] [Green Version]
- Ghareeb, M.A.; Tammam, M.A.; El-Demerdash, A.; Atanasov, A.G. Insights about Clinically Approved and Preclinically Investigated Marine Natural Products. Curr. Res. Biotechnol. 2020, 2, 88–102. [Google Scholar] [CrossRef]
- Haque, N.; Parveen, S.; Tang, T.; Wei, J.; Huang, Z. Marine Natural Products in Clinical Use. Mar. Drugs 2022, 20, 528. [Google Scholar] [CrossRef]
- Williams, P.G. Panning for Chemical Gold: Marine Bacteria as a Source of New Therapeutics. Trends Biotechnol. 2009, 27, 45–52. [Google Scholar] [CrossRef]
- Sigwart, J.D.; Blasiak, R.; Jaspars, M.; Jouffray, J.-B.; Tasdemir, D. Unlocking the Potential of Marine Biodiscovery. Nat. Prod. Rep. 2021, 38, 1235–1242. [Google Scholar] [CrossRef]
- Gago, F. Computational Approaches to Enzyme Inhibition by Marine Natural Products in the Search for New Drugs. Mar. Drugs 2023, 21, 100. [Google Scholar] [CrossRef]
- Klemetsen, T.; Raknes, I.A.; Fu, J.; Agafonov, A.; Balasundaram, S.V.; Tartari, G.; Robertsen, E.; Willassen, N.P. The MAR Databases: Development and Implementation of Databases Specific for Marine Metagenomics. Nucleic Acids Res. 2018, 46, D692–D699. [Google Scholar] [CrossRef] [PubMed]
- Lyu, C.; Chen, T.; Qiang, B.; Liu, N.; Wang, H.; Zhang, L.; Liu, Z. CMNPD: A Comprehensive Marine Natural Products Database towards Facilitating Drug Discovery from the Ocean. Nucleic Acids Res. 2021, 49, D509–D515. [Google Scholar] [CrossRef]
- MarinLit—A Database of the Marine Natural Products Literature. Available online: https://0-marinlit-rsc-org.brum.beds.ac.uk/ (accessed on 6 July 2023).
- Blin, K.; Shaw, S.; Augustijn, H.E.; Reitz, Z.L.; Biermann, F.; Alanjary, M.; Fetter, A.; Terlouw, B.R.; Metcalf, W.W.; Helfrich, E.J.N.; et al. AntiSMASH 7.0: New and Improved Predictions for Detection, Regulation, Chemical Structures and Visualisation. Nucleic Acids Res. 2023, 51, gkad344. [Google Scholar] [CrossRef]
- Sayers, E.W.; Bolton, E.E.; Brister, J.R.; Canese, K.; Chan, J.; Comeau, D.C.; Connor, R.; Funk, K.; Kelly, C.; Kim, S.; et al. Database Resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2022, 50, D20–D26. [Google Scholar] [CrossRef] [PubMed]
- Reimer, L.C.; Sardà Carbasse, J.; Koblitz, J.; Ebeling, C.; Podstawka, A.; Overmann, J. BacDive in 2022: The Knowledge Base for Standardized Bacterial and Archaeal Data. Nucleic Acids Res. 2022, 50, D741–D746. [Google Scholar] [CrossRef] [PubMed]
- Udwary, D.W.; Zeigler, L.; Asolkar, R.N.; Singan, V.; Lapidus, A.; Fenical, W.; Jensen, P.R.; Moore, B.S. Genome Sequencing Reveals Complex Secondary Metabolome in the Marine Actinomycete Salinispora Tropica. Proc. Natl. Acad. Sci. USA 2007, 104, 10376–10381. [Google Scholar] [CrossRef] [PubMed]
- Curren, E.; Leaw, C.P.; Lim, P.T.; Leong, S.C.Y. The Toxic Cosmopolitan Cyanobacteria Moorena Producens: Insights into Distribution, Ecophysiology and Toxicity. Environ. Sci. Pollut. Res. 2022, 29, 78178–78206. [Google Scholar] [CrossRef]
- Taxonomy Browser. Available online: https://0-www-ncbi-nlm-nih-gov.brum.beds.ac.uk/datasets/taxonomy/tree/?taxon=1783272,1224,1783270,1783257,200940,29547,28890,1783275 (accessed on 4 July 2023).
- Gong, X.; del Río, Á.R.; Xu, L.; Chen, Z.; Langwig, M.V.; Su, L.; Sun, M.; Huerta-Cepas, J.; De Anda, V.; Baker, B.J. New Globally Distributed Bacterial Phyla within the FCB Superphylum. Nat. Commun. 2022, 13, 7516. [Google Scholar] [CrossRef]
- Stincone, P.; Brandelli, A. Marine Bacteria as Source of Antimicrobial Compounds. Crit. Rev. Biotechnol. 2020, 40, 306–319. [Google Scholar] [CrossRef]
- Garrity, G.M.; Holt, J.G.; Hatchikian, E.C.; Ollivier, B.; Garcia, J.-L. Phylum BIII. Thermodesulfobacteria Phy. Nov. In Bergey’s Manual® of Systematic Bacteriology: Volume One: The Archaea and the Deeply Branching and Phototrophic Bacteria; Boone, D.R., Castenholz, R.W., Garrity, G.M., Eds.; Springer: New York, NY, USA, 2001; pp. 389–393. ISBN 978-0-387-21609-6. [Google Scholar]
- Wibowo, J.T.; Bayu, A.; Aryati, W.D.; Fernandes, C.; Yanuar, A.; Kijjoa, A.; Putra, M.Y. Secondary Metabolites from Marine-Derived Bacteria with Antibiotic and Antibiofilm Activities against Drug-Resistant Pathogens. Mar. Drugs 2023, 21, 50. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, R.; Kannappan, A.; Shi, C.; Lin, X. Marine Bacterial Secondary Metabolites: A Treasure House for Structurally Unique and Effective Antimicrobial Compounds. Mar. Drugs 2021, 19, 530. [Google Scholar] [CrossRef] [PubMed]
- Anburajan, L.; Meena, B.; Sreelatha, T.; Vinithkumar, N.V.; Kirubagaran, R.; Dharani, G. Ectoine Biosynthesis Genes from the Deep Sea Halophilic Eubacteria, Bacillus Clausii NIOT-DSB04: Its Molecular and Biochemical Characterization. Microb. Pathog. 2019, 136, 103693. [Google Scholar] [CrossRef] [PubMed]
- Fenizia, S.; Thume, K.; Wirgenings, M.; Pohnert, G. Ectoine from Bacterial and Algal Origin Is a Compatible Solute in Microalgae. Mar. Drugs 2020, 18, 42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galasso, C.; Corinaldesi, C.; Sansone, C. Carotenoids from Marine Organisms: Biological Functions and Industrial Applications. Antioxidants 2017, 6, 96. [Google Scholar] [CrossRef] [Green Version]
- De Mol, M.L.; Snoeck, N.; De Maeseneire, S.L.; Soetaert, W.K. Hidden Antibiotics: Where to Uncover? Biotechnol. Adv. 2018, 36, 2201–2218. [Google Scholar] [CrossRef]
- Willems, T.; De Mol, M.L.; De Bruycker, A.; De Maeseneire, S.L.; Soetaert, W.K. Alkaloids from Marine Fungi: Promising Antimicrobials. Antibiotics 2020, 9, 340. [Google Scholar] [CrossRef]
- Sadaka, C.; Ellsworth, E.; Hansen, P.R.; Ewin, R.; Damborg, P.; Watts, J.L. Review on Abyssomicins: Inhibitors of the Chorismate Pathway and Folate Biosynthesis. Molecules 2018, 23, 1371. [Google Scholar] [CrossRef] [Green Version]
- Roh, H.; Uguru, G.C.; Ko, H.-J.; Kim, S.; Kim, B.-Y.; Goodfellow, M.; Bull, A.T.; Kim, K.H.; Bibb, M.J.; Choi, I.-G.; et al. Genome Sequence of the Abyssomicin- and Proximicin-Producing Marine Actinomycete Verrucosispora Maris AB-18-032. J. Bacteriol. 2011, 193, 3391–3392. [Google Scholar] [CrossRef] [Green Version]
- Salim, A.A.; Khalil, Z.G.; Elbanna, A.H.; Wu, T.; Capon, R.J. Methods in Microbial Biodiscovery. Mar. Drugs 2021, 19, 503. [Google Scholar] [CrossRef]
- Shi, S.; Cui, L.; Zhang, K.; Zeng, Q.; Li, Q.; Ma, L.; Long, L.; Tian, X. Streptomyces Marincola Sp. Nov., a Novel Marine Actinomycete, and Its Biosynthetic Potential of Bioactive Natural Products. Front. Microbiol. 2022, 13, 860308. [Google Scholar] [CrossRef] [PubMed]
- He, H.-Y.; Pan, H.-X.; Wu, L.-F.; Zhang, B.-B.; Chai, H.-B.; Liu, W.; Tang, G.-L. Quartromicin Biosynthesis: Two Alternative Polyketide Chains Produced by One Polyketide Synthase Assembly Line. Chem. Biol. 2012, 19, 1313–1323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feling, R.H.; Buchanan, G.O.; Mincer, T.J.; Kauffman, C.A.; Jensen, P.R.; Fenical, W. Salinosporamide A: A Highly Cytotoxic Proteasome Inhibitor from a Novel Microbial Source, a Marine Bacterium of the New Genus Salinospora. Angew. Chem. Int. Ed. 2003, 42, 355–357. [Google Scholar] [CrossRef]
- Montuori, E.; Capalbo, A.; Lauritano, C. Marine Compounds for Melanoma Treatment and Prevention. Int. J. Mol. Sci. 2022, 23, 10284. [Google Scholar] [CrossRef] [PubMed]
- Atanasov, A.G.; Zotchev, S.B.; Dirsch, V.M.; Supuran, C.T. Natural Products in Drug Discovery: Advances and Opportunities. Nat. Rev. Drug Discov. 2021, 20, 200–216. [Google Scholar] [CrossRef]
- Palaniappan, K.; Chen, I.-M.A.; Chu, K.; Ratner, A.; Seshadri, R.; Kyrpides, N.C.; Ivanova, N.N.; Mouncey, N.J. IMG-ABC v.5.0: An Update to the IMG/Atlas of Biosynthetic Gene Clusters Knowledgebase. Nucleic Acids Res. 2020, 48, D422–D430. [Google Scholar] [CrossRef] [PubMed]
- Vallenet, D.; Calteau, A.; Dubois, M.; Amours, P.; Bazin, A.; Beuvin, M.; Burlot, L.; Bussell, X.; Fouteau, S.; Gautreau, G.; et al. MicroScope: An Integrated Platform for the Annotation and Exploration of Microbial Gene Functions through Genomic, Pangenomic and Metabolic Comparative Analysis. Nucleic Acids Res. 2020, 48, D579–D589. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Cumulative BLAST Score for … | ||||
|---|---|---|---|---|
| Species | … Abyssomicin | … Best Hit | Coverage Best Hit | Compound Prediction |
| Micromonospora maris | 38,594 | 38,594 | 96% | abyssomicin C |
| Streptomyces buecherae | 38,260 | 161,434 | 106% | nigericin |
| Saccharomonospora piscinae | 31,152 | 241,330 | 56% | mediomycin A |
| Streptomyces marincola | 25,243 | 120,655 | 100% | heronamide |
| Actinoalloteichus fjordicus | 17,602 | 79,117 | 16% | mediomycin A |
| Amycolatopsis albispora | 14,902 | 35,415 | 75% | quartromicin A1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hermans, C.; De Mol, M.L.; Mispelaere, M.; De Rop, A.-S.; Rombaut, J.; Nusayr, T.; Creamer, R.; De Maeseneire, S.L.; Soetaert, W.K.; Hulpiau, P. MariClus: Your One-Stop Platform for Information on Marine Natural Products, Their Gene Clusters and Producing Organisms. Mar. Drugs 2023, 21, 449. https://0-doi-org.brum.beds.ac.uk/10.3390/md21080449
Hermans C, De Mol ML, Mispelaere M, De Rop A-S, Rombaut J, Nusayr T, Creamer R, De Maeseneire SL, Soetaert WK, Hulpiau P. MariClus: Your One-Stop Platform for Information on Marine Natural Products, Their Gene Clusters and Producing Organisms. Marine Drugs. 2023; 21(8):449. https://0-doi-org.brum.beds.ac.uk/10.3390/md21080449
Chicago/Turabian StyleHermans, Cedric, Maarten Lieven De Mol, Marieke Mispelaere, Anne-Sofie De Rop, Jeltien Rombaut, Tesneem Nusayr, Rebecca Creamer, Sofie L. De Maeseneire, Wim K. Soetaert, and Paco Hulpiau. 2023. "MariClus: Your One-Stop Platform for Information on Marine Natural Products, Their Gene Clusters and Producing Organisms" Marine Drugs 21, no. 8: 449. https://0-doi-org.brum.beds.ac.uk/10.3390/md21080449