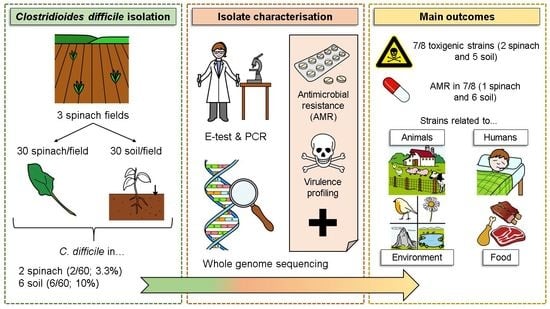
Detection and Genomic Characterisation of Clostridioides difficile from Spinach Fields
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. C. difficile Isolation and Confirmation
2.3. Characterization of Toxin and Accessory Genes
2.4. Antimicrobial Susceptibility Testing
2.5. PCR-Ribotyping
2.6. Whole Genome Sequencing (WGS)
3. Results
3.1. Characterisation of the C. difficile Isolates Using Conventional PCR, E-Test and PCR-Ribotyping
| Collection Field | Sample Type | ID 1 | RT 2 | Antibiotic Resistance Profile (MIC) | Toxin Genes 3 | Accessory Genes 3 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ERY | MET | CLIN | MOX | VAN | RIF | tcdA | tcdB | cdtA | cdtB | tcdC | tcdR | ||||
| Field 1 | Soil | 1 | 614 | S (0.25) | S (0.125) | S (8) | S (1) | S (0.75) | R (0.25) | + | + | − | − | + | + |
| Spinach | 2 | 078 | S (0.25) | S (<0.016) | S (<0.016) | S (0.38) | S (1.5) | S (<0.002) | − | + | + | + | + | + | |
| 3 | 126 | R (>256) | S (0.094) | S (4) | S (0.75) | S (1.5) | S (0.003) | − | + | + | + | + | + | ||
| Field 2 | Soil | 4 | 003 | R (8) | S (0.25) | S (1.5) | S (1) | R (>256) | R (0.125) | + | + | − | − | + | + |
| Field 3 | Soil | 5 | 087 | S (0.25) | S (0.125) | S (<0.016) | R (4) | S (1.5) | R (0.047) | + | + | − | − | + | + |
| Soil | 6 | 050 | R (4) | S (0.094) | S (1.5) | S (0.5) | R (3) | R (0.25) | + | + | − | − | + | + | |
| Soil | 7 | 511 | S (0.75) | S (0.094) | S (3) | S (0.75) | S (1.5) | R (0.19) | − | − | − | − | − | − | |
| Soil | 8 | 014/0 | S (<0.016) | S (<0.016) | R (16) | S (0.5) | S (1) | S (<0.002) | + | + | − | − | + | + | |
3.2. Characterisation of the C. difficile Isolates by Whole Genome Sequencing (WGS)
| ID 1 | AMR Using ResFinder and Prokka | AMR Using CARD | ||
|---|---|---|---|---|
| Gene | Confers Resistance to | Gene | Confers Resistance to | |
| 1 | vanB2 | Vancomycin | vanXY gene in vanG cluster | Vancomycin |
| vanR gene in vanG cluster | ||||
| ND 3 | CDD-1 | Beta-lactams | ||
| ND 3 | qacG | Disinfecting agents and antiseptics | ||
| ND 3 | Clostridioides difficile 23S rRNA mutation | Erythromycin and clindamycin | ||
| 2 | tet(M)_10 | Tetracyclines | tet(M) | Tetracyclines |
| ant(6)-Ia_2 | Aminoglycosides | ant(6)-Ia | Aminoglycosides | |
| ND 3 | CDD-1 | Beta-lactams | ||
| ND 3 | qacG | Disinfecting agents and antiseptics | ||
| ND 3 | Clostridioides difficile 23S rRNA mutation | Erythromycin and clindamycin | ||
| 3 | D19aph(3′)-III_1 | Aminoglycosides | APH(3′)-IIIa | Aminoglycosides |
| ant(6)-Ia_1 | aad(6) | |||
| ant(6)-Ia_3 | ||||
| tet(40)_1 | Tetracyclines | tet(40) | Tetracyclines | |
| tet(M)_4 | ||||
| ND 3 | CDD-1 | Beta-lactams | ||
| ND 3 | SAT-4 | Nucleosides | ||
| ND 3 | qacG | Disinfecting agents and antiseptics | ||
| ND 3 | Clostridioides difficile 23S rRNA mutation | Erythromycin and clindamycin | ||
| 4 | vanB 2 | Vancomycin | vanXY gene in vanG cluster | Vancomycin |
| vanR gene in vanG cluster | ||||
| ND 3 | CDD-2 | Beta-lactams | ||
| ND 3 | qacG | Disinfecting agents and antiseptics | ||
| ND 3 | Clostridioides difficile 23S rRNA mutation | Erythromycin and clindamycin | ||
| 5 | vanB 2 | Vancomycin | vanXY gene in vanG cluster | Vancomycin |
| vanR gene in vanG cluster | ||||
| ND 3 | CDD-1 | Beta-lactams | ||
| ND 3 | cdeA | Fluoroquinolones, disinfecting agents and antiseptics | ||
| ND 3 | qacG | Disinfecting agents and antiseptics | ||
| ND 3 | Clostridioides difficile 23S rRNA mutation | Erythromycin and clindamycin | ||
| 6 | vanB 2 | Vancomycin | vanXY gene in vanG cluster | Vancomycin |
| vanR gene in vanG cluster | ||||
| ND 3 | CDD-1 | Beta-lactams | ||
| ND 3 | qacG | Disinfecting agents and antiseptics | ||
| ND 3 | Clostridioides difficile 23S rRNA mutation | Erythromycin and clindamycin | ||
| 7 | ND 3 | vanXY gene in vanG cluster | Vancomycin | |
| vanR gene in vanG cluster | ||||
| ND 3 | CDD-1 | Beta-lactams | ||
| ND 3 | qacG | Disinfecting agents and antiseptics | ||
| ND 3 | Clostridioides difficile 23S rRNA mutation | Erythromycin and clindamycin | ||
| 8 | ND 3 | vanXY gene in vanG cluster | Vancomycin | |
| vanR gene in vanG cluster | ||||
| ND 3 | CDD-1 | Beta-lactams | ||
| ND 3 | qacG | Disinfecting agents and antiseptics | ||
| ND 3 | Clostridioides difficile 23S rRNA mutation | Erythromycin and clindamycin | ||
4. Discussion
4.1. Prevalence of C. difficile
4.2. Ribotypes of C. difficile
4.3. Toxigenicity of the C. difficile Isolates
4.4. Antibiotic Susceptibility of the C. difficile Isolates
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Enoch, D.A.; Aliyu, S.H. Is Clostridium difficile infection still a problem for hospitals? Can. Med. Assoc. J. 2012, 184, 17–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Czepiel, J.; Dróżdż, M.; Pituch, H.; Kuijper, E.J.; Perucki, W.; Mielimonka, A.; Goldman, S.; Wultańska, D.; Garlicki, A.; Biesiada, G. Clostridium difficile infection: Review. Eur. J. Clin. Microbiol. Infect. Dis. 2019, 38, 1211–1221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, A.M.; Kuijper, E.J.; Wilcox, M.H. Clostridium difficile: A European perspective. J. Infect. 2013, 66, 115–128. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Tunis, M.C.; Frenette, C.; Katz, K.; Amaratunga, K.; Rhodenizer Rose, S.; House, A.; Quach, C. Epidemiology of Clostridioides difficile infection in Canada: A six-year review to support vaccine decision-making. CCDR 2019, 45, 191–211. [Google Scholar] [CrossRef] [PubMed]
- Donta, S.T.; Sullivan, N.; Wilkins, T.D. Differential effects of Clostridium difficile toxins on tissue-cultured cells. J. Clin. Microbiol. 1982, 15, 1157–1158. [Google Scholar] [CrossRef] [Green Version]
- Feltis, B.A.; Wiesner, S.M.; Kim, A.S.; Erlandsen, S.L.; Lyerly, D.L.; Wilkins, T.D.; Wells, C.L. Clostridium difficile toxins A and B can alter epithelial permeability and promote bacterial paracellular migration through HT-29 enterocytes. Shock 2000, 14, 629–634. [Google Scholar] [CrossRef]
- Riggs, M.M.; Sethi, A.K.; Zabarsky, T.F.; Eckstein, E.C.; Jump, R.L.P.; Donskey, C.J. Asymptomatic carriers are a potential source for transmission of epidemic and nonepidemic Clostridium difficile strains among longterm care facility residents. Clin. Infect. Dis. 2007, 45, 992–998. [Google Scholar] [CrossRef] [Green Version]
- Rupnik, M.; Janezic, S. An update on Clostridium difficile toxinotyping. J. Clin. Microbiol. 2016, 54, 13–18. [Google Scholar] [CrossRef] [Green Version]
- Voth, D.; Ballard, J.D. Clostridium difficile Toxins: Mechanism of Action and Role in Disease. Clin. Microbiol. Rev. 2005, 2, 247–263. [Google Scholar] [CrossRef] [Green Version]
- Kuehne, S.A.; Collery, M.M.; Kelly, M.L.; Cartman, S.T.; Cockayne, A.; Minton, N.P. Importance of toxin A, toxin B, and CDT in virulence of an epidemic Clostridium difficile strain. J. Infect. Dis. 2014, 209, 83–86. [Google Scholar] [CrossRef]
- Gerding, D.N.; Johnson, S.; Rupnik, M.; Aktories, K. Clostridium difficile binary toxin CDT: Mechanism, epidemiology, and potential clinical importance. Gut Microbes 2014, 5, 15–27. [Google Scholar] [CrossRef] [Green Version]
- Stiles, B.G.; Wigelsworth, D.J.; Popoff, M.R.; Barth, H. Clostridial binary toxins: Iota and c2 family portraits. Front. Cell. Infect. Microbiol. 2011, 1, 11. [Google Scholar] [CrossRef] [Green Version]
- Dingle, T.C.; MacCannell, D.R. Chapter 9—Molecular Strain Typing and Characterisation of Toxigenic Clostridium difficile. In Methods in Microbiology; Sails, A., Tang, Y.-W., Eds.; Academic Press: San Diego, CA, USA, 2015; Volume 42, pp. 329–357. [Google Scholar]
- Freeman, J.; Vernon, J.; Morris, K.; Nicholson, S.; Todhunter, S.; Longshaw, C.; Wilcox, M.H. Pan-European Longitudinal Surveillance of Antibiotic Resistance among Prevalent Clostridium difficile Ribotypes’ Study Group. Clin. Microbiol. Infect. 2015, 21, 248.e9–248.e16. [Google Scholar] [CrossRef] [Green Version]
- Hampikyan, H.; Bingol, E.B.; Muratoglu, K.; Akkaya, E.; Cetin, O.; Colak, H. The prevalence of Clostridium difficile in cattle and sheep carcasses and the antibiotic susceptibility of isolates. Meat Sci. 2018, 139, 120–124. [Google Scholar] [CrossRef]
- Herbert, R.; Hatcher, J.; Jauneikaite, E.; Gharbi, M.; d’Arc, S.; Obaray, N.; Rickards, T.; Rebec, M.; Blandy, O.; Hope, R.; et al. Two-year analysis of Clostridium difficile ribotypes associated with increased severity. J. Hosp. Infect. 2019, 103, 388–394. [Google Scholar] [CrossRef] [Green Version]
- Couturier, J.; Davies, K.; Gateau, C.; Barbut, F. Ribotypes and New Virulent Strains Across Europe. Adv. Exp. Med. Biol. 2018, 1050, 45–58. [Google Scholar]
- Peng, Z.; Jin, D.; Kim, H.B.; Stratton, C.W.; Wu, B.; Tang, Y.-W.; Sun, X. Update on Antimicrobial Resistance in Clostridium difficile: Resistance Mechanisms and Antimicrobial Susceptibility Testing. J. Clin. Microbiol. 2017, 55, 1998–2008. [Google Scholar] [CrossRef] [Green Version]
- He, M.; Miyajima, F.; Roberts, P.; Ellison, L.; Pickard, D.J.; Martin, M.J.; Connor, T.R.; Harris, S.R.; Fairley, D.; Bamford, B.; et al. Emergence and global spread of epidemic healthcare-associated Clostridium difficile. Nat. Genet. 2013, 45, 109–113. [Google Scholar] [CrossRef]
- Owens, R.C.; Donskey, C.J.; Gaynes, R.P.; Loo, V.G.; Muto, C.A. Antimicrobial-associated risk factors for Clostridium difficile infection. Clin. Infect. Dis. 2008, 46, 19–31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spigaglia, P.; Barbanti, F.; Mastrantonio, P. European Study Group on Clostridium difficile (ESGCD) Multidrug resistance in European Clostridium difficile clinical isolates. J. Antimicrob. Chemother. 2011, 66, 2227–2234. [Google Scholar] [CrossRef] [Green Version]
- Cole, S.A.; Stahl, T.J. Persistent and Recurrent Clostridium difficile Colitis. Clin. Colon Rectal Surg. 2015, 28, 65–69. [Google Scholar] [CrossRef]
- Haagsma, J. Pathogenic anaerobic bacteria and the environment. Rev. Sci. Tech. (Int. Off. Epizoot.) 1991, 10, 749–764. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marcos, P.; Whyte, P.; Rogers, T.; McElroy, M.; Fanning, S.; Frias, J.; Bolton, D. The prevalence of Clostridioides difficile on farms, in abattoirs and in retail foods in Ireland. Food Microbiol. 2021, 98, 103781. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, C.; Bouchafa, L.; Soumillion, K.; Ngyuvula, E.; Taminiau, B.; Van Broeck, J.; Delmée, M.; Daube, G. Seasonality of Clostridium difficile in the natural environment. Transbound. Emerg. Dis. 2019, 66, 2440–2449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shivaperumal, N.; Chang, B.J.; Riley, T.V. High Prevalence of Clostridium difficile in Home Gardens in Western Australia. Appl. Environ. Microbiol. J. 2020, 87, e01572-20. [Google Scholar] [CrossRef]
- Bazaid, F. Distribution and Sources of Clostridium difficile Present in Water Sources. Ph.D. Thesis, University of Guelph, Guelph, ON, Canada, 2013. [Google Scholar]
- Tkalec, V.; Janezica, S.; Skoka, B.; Simonica, T.; Mesarica, S.; Vrabica, T.; Rupnik, M. High Clostridium difficile contamination rates of domestic and imported potatoes compared to some other vegetables in Slovenia. Food Microbiol. 2019, 78, 194–200. [Google Scholar] [CrossRef]
- Warriner, K.; Xu, C.; Habash, M.; Sultan, S.; Weese, S.J. Dissemination of Clostridium difficile in food and the environment: Significant sources of, C. difficile community acquired infection? J. Appl. Microbiol. 2016, 122, 542–555. [Google Scholar] [CrossRef]
- Xu, C.; Weese, J.S.; Flemming, C.; Odumeru, J.; Warriner, K. Fate of Clostridium difficile during wastewater treatment and incidence in Southern Ontario watersheds. J. Appl. Microbiol. 2014, 117, 891–904. [Google Scholar] [CrossRef]
- Eckert, C.; Burghoffer, B.; Barbut, F. Contamination of ready-to-eat raw vegetables with Clostridium difficile, France. J. Med. Microbiol. 2013, 62, 1435–1438. [Google Scholar] [CrossRef]
- Lim, S.C.; Foster, N.F.; Elliott, B.; Riley, T.V. High prevalence of Clostridium difficile on retail root vegetables, Western Australia. J. Appl. Microbiol. 2017, 124, 585–590. [Google Scholar] [CrossRef]
- Metcalf, D.; Costa, M.C.; Dew, W.M.; Weese, J.S. Clostridium difficile in vegetables, Canada. Lett. Appl. Microbiol. 2010, 51, 600–602. [Google Scholar] [CrossRef]
- Primavilla, S.; Farneti, S.; Petruzzelli, A.; Drigo, I.; Scuota, S. Contamination of hospital food with Clostridium difficile in Central Italy. Anaerobe 2019, 55, 8–10. [Google Scholar] [CrossRef]
- Bakri, M.M.; Brown, D.J.; Butcher, J.P.; Sutherland, A.D. Clostridium difficile in ready-to-eat salads, Scotland. Emerg. Infect. Dis. 2009, 15, 817–818. [Google Scholar] [CrossRef]
- Yamoudy, M.; Mirlohi, M.; Isfahani, B.N.; Jalali, M.; Esfandiari, Z.; Hosseini, N.S. Isolation of toxigenic Clostridium difficile from ready-to-eat salads by multiplex polymerase chain reaction in Isfahan, Iran. Adv. Biomed. Res. 2015, 4, 87. [Google Scholar]
- Lim, S.C.; Knight, D.R.; Moono, P.; Foster, N.F.; Riley, T.V. Clostridium difficile in soil conditioners, mulches and garden mixes with evidence of a clonal relationship with historical food and clinical isolates. Environ. Microbiol. Rep. 2020, 12, 672–680. [Google Scholar] [CrossRef]
- Le Maréchal, C.; Druilhe, C.; Repérant, E.; Boscher, E.; Rouxel, S.; Le Roux, S.; Poëzévara, T.; Ziebal, C.; Houdayer, C.; Nagard, B.; et al. Evaluation of the occurrence of sporulating and nonsporulating pathogenic bacteria in manure and in digestate of five agricultural biogas plants. Microbiol. Open 2019, 8, e872. [Google Scholar] [CrossRef]
- Lemee, L.; Dhalluin, A.; Testelin, S.; Mattrat, M.A.; Maillard, K.; Lemeland, J.F.; Pons, J.L. Multiplex PCR targeting tpi (triose phosphate isomerase), tcdA (toxin A), and tcdB (toxin B) genes for toxigenic culture of Clostridium difficile. J. Clin. Microbiol. 2004, 42, 5710–5714. [Google Scholar] [CrossRef] [Green Version]
- Ackermann, G.; Tang-Feldman, Y.J.; Schaumann, R.; Henderson, J.P.; Rodloff, A.C.; Silva, J.; Cohen, S.H. Antecedent use of fluoroquinolones is associated with resistance to moxifloxacin in Clostridium difficile. Clin. Microbiol. Infect. 2003, 9, 526–530. [Google Scholar] [CrossRef] [Green Version]
- Oldfield, E.C., IV; Oldfield, E.C., III; Johnson, D.A. Clinical update for the diagnosis and treatment of Clostridium difficile infection. World J. Gastrointest. Pharmacol. Ther. 2014, 5, 1–26. [Google Scholar]
- Johnson, S.; Samore, M.H.; Farrow, K.A.; Killgore, G.E.; Tenover, F.C.; Lyras, D.; Rood, J.I.; DeGirolami, P.; Baltch, A.L.; Rafferty, M.E.; et al. Epidemics of diarrhea caused by a clindamycin-resistant strain of Clostridium difficile in four hospitals. N. Engl. J. Med. 1999, 341, 1645–1651. [Google Scholar] [CrossRef]
- Surawicz, C.M.; Brandt, L.J.; Binion, D.G.; Ananthakrishnan, A.N.; Curry, S.R.; Gilligan, P.H.; McFarland, L.V.; Mellow, M.; Zuckerbraun, B.S. Guidelines for diagnosis, treatment, and prevention of Clostridium difficile infections. Am. J. Gastroenterol. 2013, 108, 478–498. [Google Scholar] [CrossRef] [PubMed]
- Vardakas, K.Z.; Polyzos, K.A.; Patouni, K.; Rafailidis, P.I.; Samonis, G.; Falagas, M.E. Treatment failure and recurrence of Clostridium difficile infection following treatment with vancomycin or metronidazole: A systematic review of the evidence. Int. J. Antimicrob. Agents 2012, 40, 1–8. [Google Scholar] [CrossRef] [PubMed]
- European Committee on Antimicrobial Susceptibility Testing (EUCAST). Breakpoint Tables for Interpretation of MICs and Zone Diameters Version 11.0, Valid from 2021-01-01. 2021. Available online: http://www.eucast.org (accessed on 23 November 2021).
- European Centre for Disease Prevention and Control (ECDC). Laboratory Procedures for Diagnosis and Typing of Human Clostridium difficile Infection. Technical Report. 2018. Available online: www.ecdc.europa.eu (accessed on 23 November 2021).
- Bidet, P.; Barbut, F.; Lalande, V.; Burghoffer, B.; Petit, J.C. Development of a new PCR-ribotyping method for Clostridium difficile based on ribosomal RNA gene sequencing. FEMS Microbiol. Lett. 1999, 175, 261–266. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data [Online]. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 20 May 2022).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wood, D.E.; Lu, J.; Langmead, B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019, 20, 257. [Google Scholar] [CrossRef] [Green Version]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [Green Version]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef] [Green Version]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [Green Version]
- Clausen, P.T.L.C.; Aarestrup, F.M.; Lund, O. Rapid and precise alignment of raw reads against redundant databases with KMA. BMC Bioinform. 2018, 19, 307. [Google Scholar] [CrossRef]
- Hasman, H.; Saputra, D.; Sicheritz-Ponten, T.; Lund, O.; Svendsen, C.A.; Frimodt-Møller, N.; Aarestrup, F.M. Rapid whole-genome sequencing for detection and characterization of microorganisms directly from clinical samples. J. Clin. Microbiol. 2014, 52, 139–146. [Google Scholar] [CrossRef] [Green Version]
- Larsen, M.V.; Cosentino, S.; Lukjancenko, O.; Saputra, D.; Rasmussen, S.; Hasman, H.; Sicheritz-Ponten, T.; Aarestrup, F.M.; Ussery, D.W.; Lund, O. Benchmarking of methods for genomic taxonomy. J. Clin. Microbiol. 2014, 52, 1529–1539. [Google Scholar] [CrossRef] [Green Version]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Zankari, E.; Allesøe, R.; Joensen, K.G.; Cavaco, L.M.; Lund, O.; Aarestrup, F.M. PointFinder: A novel web tool for WGS-based detection of antimicrobial resistance associated with chromosomal point mutations in bacterial pathogens. J. Antimicrob. Chemother. 2020, 72, 2764–2768. [Google Scholar] [CrossRef] [Green Version]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef]
- McArthur, A.G.; Waglechner, N.; Nizam, F.; Yan, A.; Azad, M.A.; Baylay, A.J.; Bhullar, K.; Canova, M.J.; De Pascale, G.; Ejim, L.; et al. The comprehensive antibiotic resistance database. Antimicrob. Agents Chemother. 2013, 57, 3348–3357. [Google Scholar] [CrossRef] [Green Version]
- Larsen, M.V.; Cosentino, S.; Rasmussen, S.; Friis, C.; Hasman, H.; Marvig, R.L.; Jelsbak, L.; Sicheritz-Pontén, T.; Ussery, D.W.; Aarestrup, F.M.; et al. Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 2012, 50, 1355–1361. [Google Scholar] [CrossRef] [Green Version]
- Seemann, T. Snippy: Fast Bacterial Variant Calling from NGS Reads. 2015. Available online: https://github.com/tseemann/snippy (accessed on 1 June 2022).
- Eren, A.M.; Kiefl, E.; Shaiber, A.; Veseli, I.; Miller, S.E.; Schechter, M.S.; Fink, I.; Pan, J.N.; Yousef, M.; Fogarty, E.C.; et al. Community-led, integrated, reproducible multi-omics with anvi’o. Nat. Microbiol. 2021, 6, 3–6. [Google Scholar] [CrossRef]
- Hyatt, D.; Chen, G.L.; Locascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef] [Green Version]
- Tatusov, R.L.; Galperin, M.Y.; Natale, D.A.; Koonin, E.V. The COG database: A tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000, 28, 33–36. [Google Scholar] [CrossRef] [Green Version]
- Benedict, M.N.; Henriksen, J.R.; Metcalf, W.W.; Whitaker, R.J.; Price, N.D. ITEP: An integrated toolkit for exploration of microbial pan-genomes. BMC Genom. 2014, 15, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Dongen, S.; Abreu-Goodger, C. Using MCL to extract clusters from networks. Methods Mol. Biol. 2012, 804, 281–295. [Google Scholar] [PubMed]
- Leekitcharoenphon, P.; Nielsen, E.M.; Kaas, R.S.; Lund, O.; Aarestrup, F.M. Evaluation of whole genome sequencing for outbreak detection of Salmonella enterica. PLoS ONE 2014, 9, e87991. [Google Scholar] [CrossRef] [PubMed]
- Joensen, K.G.; Scheutz, F.; Lund, O.; Hasman, H.; Kaas, R.S.; Nielsen, E.M.; Aarestrup, F.M. Real-Time Whole-Genome Sequencing for Routine Typing, Surveillance, and Outbreak Detection of Verotoxigenic Escherichia coli. J. Clin. Microbiol. 2014, 52, 1501–1510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaas, R.S.; Leekitcharoenphon, P.; Aarestrup, F.M.; Lund, O. Solving the Problem of Comparing Whole Bacterial Genomes across Different Sequencing Platforms. PLoS ONE 2014, 9, e104984. [Google Scholar] [CrossRef] [Green Version]
- Olaitan, A.; Dureja, C.; Youngblom, M.; Topf, M.; Shen, W.-J.; Gonzales-Luna, A.; Deshpande, A.; Hevener, K.; Freeman, J.; Wilcox, M.; et al. Decoding a cryptic mechanism of metronidazole resistance among globally disseminated fluoroquinolone-resistant Clostridioides difficile. bioRxiv 2022. [Google Scholar] [CrossRef]
- Dang, U.T.; Zamora, I.; Hevener, K.E.; Adhikari, S.; Wu, X.; Hurdle, J.G. Rifamycin Resistance in Clostridium difficile Is Generally Associated with a Low Fitness Burden. Antimicrob. Agents Chemother. 2016, 60, 5604–5607. [Google Scholar] [CrossRef] [Green Version]
- Al Saif, N.; Brazier, J.S. The distribution of Clostridium difficile in the environment of South Wales. J. Med. Microbiol. 1996, 45, 133–137. [Google Scholar] [CrossRef] [Green Version]
- Simango, C. Prevalence of Clostridium difficile in the environment in a rural community in Zimbabwe. R. Soc. Trop. Med. Hyg. 2006, 100, 1146–1150. [Google Scholar] [CrossRef]
- Blasi, F.; Lovito, C.; Albini, E.; Bano, L.; Dalmonte, G.; Drigo, I.; Maresca, C.; Massacci, F.R.; Orsini, S.; Primavilla, S.; et al. Clostridioides difficile in Calves in Central Italy: Prevalence, Molecular Typing, Antimicrobial Susceptibility and Association with Antibiotic Administration. Animals 2021, 11, 515. [Google Scholar] [CrossRef]
- Hammitt, M.C.; Bueschel, D.M.; Keel, M.K.; Glock, R.D.; Cuneo, P.; DeYoung, D.W.; Reggiardo, C.; Trinh, H.T.; Songer, J.G. A possible role for Clostridium difficile in the etiology of calf enteritis. Vet. Microbiol. 2008, 127, 343–352. [Google Scholar] [CrossRef]
- Magistrali, C.F.; Maresca, C.; Cucco, L.; Bano, L.; Drigo, I.; Filippini, G.; Dettori, A.; Broccatelli, S.; Pezzotti, G. Prevalence and risk factors associated with Clostridium difficile shedding in veal calves in Italy. Anaerobe 2015, 33, 42–47. [Google Scholar] [CrossRef]
- Harvey, R.; Norman, K.; Andrews, K.; Hume, M.; Scanlan, C.; Callaway, T. Clostridium difficile in poultry and poultry meat. Foodborne Pathog. Dis. 2011, 8, 1321–1323. [Google Scholar] [CrossRef]
- Hussain, I.; Borah, P.; Sharma, R.K.; Rajkhowa, S.; Rupnik, M.; Saikia, D. P Molecular characteristics of Clostridium difficile isolates from human and animals in the North Eastern region of India. Mol. Cell. Probes 2016, 30, 306–311. [Google Scholar] [CrossRef]
- Álvarez-Pérez, S.; Blanco, J.L.; Peláez, T.; Astorga, R.J.; Harmanus, C.; Kuijper, E.; García, M.E. High prevalence of the epidemic Clostridium difficile PCR ribotype 078 in Iberian free-range pigs. Res. Vet. Sci. 2013, 95, 358–361. [Google Scholar] [CrossRef]
- Knight, D.R.; Elliott, B.; Chang, B.J.; Perkins, T.T.; Riley, T.V. Diversity and evolution in the genome of Clostridium difficile. Clin. Microbiol. Rev. 2015, 28, 721–741. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.-J.; Yang, L.; Gu, X.-X.; Chen, P.-X.; Fu, J.-L.; Jiang, H.-X. The first isolation of Clostridium difficile RT078/ST11 from pigs in China. PLoS ONE 2019, 14, e0212965. [Google Scholar]
- Debast, S.B.; van Leengoed, L.A.; Goorhuis, A.; Harmanus, C.; Kuijper, E.J.; Bergwerff, A.A. Clostridium difficile PCR ribotype 078 toxinotype, V. found in diarrhoeal pigs identical to isolates from affected humans. Environ. Microbiol. 2009, 11, 505–511. [Google Scholar] [CrossRef]
- Keessen, E.C.; Harmanus, C.; Dohmen, W.; Kuijper, E.J.; Lipman, L.J. Clostridium difficile infection associated with pig farms. Emerg. Infect. Dis. 2013, 19, 1032–1034. [Google Scholar] [CrossRef]
- Songer, J.G.; Trinh, H.T.; Killgore, G.E.; Thompson, A.D.; McDonald, L.C.; Limbago, B.M. Clostridium difficile in Retail Meat Products, USA, 2007. Emerg. Infect. Dis. 2009, 15, 819–821. [Google Scholar] [CrossRef]
- Weese, J.; Avery, B.P.; Rousseau, J.; Reid-Smith, J. Detection and Enumeration of Clostridium difficile Spores in Retail Beef and Pork. Appl. Environ. Microbiol. 2009, 15, 5009–5011. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Health Service Executive (HSE)—Health Protection Surveillance Centre (HPSC). Enhanced Surveillance of Clostridioides (Clostridium) difficile Infection in Ireland: Q2 2021 National Report; HSE-HPSC: Dublin, Ireland, 2021. [Google Scholar]
- Rabold, D.; Espelage, W.; Abu Sin, M.; Eckmanns, T.; Schneeberg, A.; Neubauer, H.; Möbius, N.; Hille, K.; Wieler, L.H.; Seyboldt, C.; et al. The zoonotic potential of Clostridium difficile from small companion animals and their owners. PLoS ONE 2018, 13, e0193411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, A.; Davenport, K.W.; Vuyisich, G.; Kunde, Y.A.; Johnson, S.L.; Chain, P.S.G.; Dichosa, A.E.K.; Rodriguez-Palacios, A. Complete Genome Sequences of Historic Clostridioides difficile Food-Dwelling Ribotype 078 Strains in Canada Identical to That of the Historic Human Clinical Strain M120 in the United Kingdom. Microbiol. Resour. Announc. 2018, 7, e00853-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martín-Burriel, I.; Andrés-Lasheras, S.; Harders, F.; Mainar-Jaime, R.C.; Ranera, B.; Zaragoza, P.; Falceto, V.; Bolea, Y.; Kuijper, E.; Bolea, R.; et al. Molecular analysis of three Clostridium difficile strain genomes isolated from pig farm-related samples. Anaerobe 2017, 48, 224–231. [Google Scholar] [CrossRef] [PubMed]
- Knight, D.R.; Putsathit, P.; Elliott, B.; Riley, T.V. Contamination of Australian newborn calf carcasses at slaughter with Clostridium difficile. Clin. Microbiol. Infect. 2016, 22, 266.e1–266.e7. [Google Scholar] [CrossRef] [Green Version]
- Masarikova, M.; Simkova, I.; Plesko, M.; Eretova, V.; Krutova, M.; Cizek, A. The Colonisation of Calves in Czech Large-Scale Dairy Farms by Clonally-Related Clostridioides difficile of the Sequence Type 11 Represented by Ribotypes 033 and 126. Microorganisms 2020, 8, 901. [Google Scholar] [CrossRef]
- Avberšek, J.; Janezic, S.; Pate, M.; Rupnik, M.; Zidaric, V.; Logar, K.; Vengust, M.; Zemljic, M.; Pirs, T.; Ocepek, M. Diversity of Clostridium difficile in pigs and other animals in Slovenia. Anaerobe 2009, 15, 252–255. [Google Scholar] [CrossRef]
- Andrés-Lasheras, S.; Bolea, R.; Mainar-Jaime, R.C.; Kuijper, E.; Sevilla, E.; Martín-Burriel, I.; Chirino-Trejo, M. Presence of Clostridium difficile in pig faecal samples and wild animal species associated with pig farms. J. Appl. Microbiol. 2017, 122, 462–472. [Google Scholar] [CrossRef]
- Agnoletti, F.; Arcangelia, G.; Barbantib, F.; Barcoa, L.; Brunettaa, R.; Cocchia, M.; Conederaa, G.; D’Estea, L.; Drigoa, I.; Spigagliab, P.; et al. Survey, characterization and antimicrobial susceptibility of Clostridium difficile from marine bivalve shellfish of North Adriatic Sea. Int. J. Food Microbiol. 2019, 298, 74–80. [Google Scholar] [CrossRef]
- Troiano, T.; Harmanus, C.; Sanders, I.; Pasquale, V.; Dumontet, S.; Capuano, F.; Romano, V.; Kuijper, E. J Toxigenic Clostridium difficile PCR ribotypes in ediblemarine bivalve molluscs in Italy. Int. J. Food Microbiol. 2015, 208, 30–34. [Google Scholar] [CrossRef]
- Álvarez-Pérez, S.; Blanco, J.L.; Harmanus, C.; Kuijper, E.; García, M.E. Subtyping and antimicrobial susceptibility of Clostridium difficile PCR ribotype 078/126 isolates of human and animal origin. Vet. Microbiol. 2017, 199, 15–22. [Google Scholar] [CrossRef]
- Santos, A.; Isidro, J.; Silva, C.; Boaventura, L.; Diogo, J.; Faustino, A.; Toscano, C.; Oleastro, M. Molecular and epidemiologic study of Clostridium difficile reveals unusual heterogeneity in clinical strains circulating in different regions in Portugal. Clin. Microbiol. Infect. 2016, 22, 695–700. [Google Scholar] [CrossRef] [Green Version]
- Azimirad, M.; Krutova, M.; Yadegar, A.; Shahrokh, S.; Olfatifar, M.; Aghdaei, H.A.; Fawley, W.N.; Wilcox, M.H.; Zali, M.R. Clostridioides difficile ribotypes 001 and 126 were predominant in Tehran healthcare settings from 2004 to 2018: A 14-year-long cross-sectional study. Emerg. Microbes Infect. 2020, 9, 1432–1443. [Google Scholar] [CrossRef]
- Hung, Y.P.; Lin, H.J.; Tsai, B.Y.; Liu, H.C.; Liu, H.C.; Lee, J.C.; Wu, Y.H.; Wilcox, M.H.; Fawley, W.N.; Hsueh, P.R.; et al. Clostridium difficile ribotype 126 in southern Taiwan: A cluster of three symptomatic cases. Anaerobe 2014, 30, 188–192. [Google Scholar] [CrossRef]
- Jamal, W.Y.; Rotimi, V.O. Surveillance of antibiotic resistance among hospital- and community-acquired toxigenic Clostridium difficile isolates over 5-year period in Kuwait. PLoS ONE 2016, 11, e0161411. [Google Scholar] [CrossRef] [Green Version]
- Ahn, S.W.; Lee, S.H.; Kim, U.J.; Jang, H.C.; Choi, H.J.; Choy, H.E.; Kang, S.J.; Roh, S.W. Genomic characterization of nine Clostridioides difficile strains isolated from Korean patients with Clostridioides difficile infection. Gut Pathog. 2021, 13, 55. [Google Scholar] [CrossRef]
- Su, T.; Chen, W.; Wang, D.; Cui, Y.; Ni, Q.; Jiang, C.; Dong, D.; Peng, Y. Complete Genome Sequencing and Comparative Phenotypic Analysis Reveal the Discrepancy Between Clostridioides difficile ST81 and ST37 Isolates. Front. Microbiol. 2021, 12, 776892. [Google Scholar] [CrossRef]
- Koene, M.; Mevius, D.; Wagenaar, J.; Harmanus, C.; Hensgens, M.; Meetsma, A.; Putirulan, F.; van Bergen, M.; Kuijper, E. Clostridium difficile in Dutch animals: Their presence, characteristics and similarities with human isolates. Clin. Microbiol. Infect. 2012, 18, 778–784. [Google Scholar] [CrossRef] [Green Version]
- de Boer, E.; Zwartkruis-Nahuis, A.; Heuvelink, A.E.; Harmanus, C.; Kuijper, E.J. Prevalence of Clostridium difficile in retailed meat in the Netherlands. Int. J. Food Microbiol. 2011, 144, 561–564. [Google Scholar] [CrossRef]
- Kecerova, Z.; Cizek, A.; Nyc, O.; Krutova, M. Clostridium difficile isolates derived from Czech horses are resistant to enrofloxacin; cluster to clades 1 and 5 and ribotype 033 predominates. Anaerobe 2019, 56, 17–21. [Google Scholar] [CrossRef]
- Yin, C.; Chen, D.S.; Zhuge, J.; McKenna, D.; Sagurton, J.; Wang, G.; Huang, W.; Dimitrova, N.; Fallon, J.T. Complete Genome Sequences of Four Toxigenic Clostridium difficile Clinical Isolates from Patients of the Lower Hudson Valley, New York, USA. Genome Announc. 2018, 6, e01537-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tkalec, V.; Jamnikar-Ciglenecki, U.; Rupnik, M.; Vadnjal, S.; Zelenik, K.; Biasizzo, M. Clostridioides difficile in national food surveillance, Slovenia, 2015 to 2017. Euro Surveill. 2020, 25, 1900479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stein, K.; Egan, S.; Lynch, H.; Harmanus, C.; Kyne, L.; Herra, C.; McDermott, S.; Kuijper, E.; Fitzpatrick, F.; FitzGerald, S.; et al. PCR-ribotype distribution of Clostridium difficile in Irish pigs. Anaerobe 2017, 48, 237–241. [Google Scholar] [CrossRef] [PubMed]
- Aguilar-Zamora, E.; Weimer, B.C.; Torres, R.C.; Gómez-Delgado, A.; Ortiz-Olvera, N.; Aparicio-Ozores, G.; Barbero-Becerra, V.J.; Torres, J.; Camorlinga-Ponce, M. Molecular Epidemiology and Antimicrobial Resistance of Clostridioides difficile in Hospitalized Patients From Mexico. Front. Microbiol. 2022, 12, 787451. [Google Scholar] [CrossRef] [PubMed]
- van Rossen, T.M.; van Prehn, J.; Koek, A.; Jonges, M.; van Houdt, R.; van Mansfeld, R.; Kuijper, E.J.; Vandenbroucke-Grauls, C.M.J.E.; Budding, A.E. Simultaneous detection and ribotyping of Clostridioides difficile, and toxin gene detection directly on fecal samples. Antimicrob. Resist. Infect. Control 2021, 10, 23. [Google Scholar] [CrossRef]
- National Centre for Biotechnology Information (NCBI). Clostridioides difficile Strain CFSAN096664 Chromosome, Complete Genome; NCBI Reference Sequence: NZ_CP049958.1. 2020. Available online: https://0-www-ncbi-nlm-nih-gov.brum.beds.ac.uk/genome/535?genome_assembly_id=1483731 (accessed on 1 August 2022).
- Rodriguez-Palacios, A.; Staempfli, H.R.; Duffield, T.; Weese, J.S. Clostridium difficile in retail ground meat, Canada. Emerg. Infect. Dis. 2007, 13, 485–487. [Google Scholar] [CrossRef]
- Schwanbeck, J.; Riedel, T.; Laukien, F.; Schober, I.; Oehmig, I.; Zimmermann, O.; Overmann, J.; Groß, U.; Zautner, A.E.; Bohne, W. Characterization of a clinical Clostridioides difficile isolate with markedly reduced fidaxomicin susceptibility and a V1143D mutation in rpoB. J. Antimicrob. Chemother. 2019, 74, 6–10. [Google Scholar] [CrossRef]
- Nazareth, C.; Leitão, I.; Reis, E.; Inácio, H.; Martins, F.; Ramalheira, E.; Cunha, F.; Santos, C.; Lino, S.; Moreira, H.; et al. Epidemiology of Clostridioides difficile Infection in Portugal: A Retrospective, Observational Study of Hospitalized Patients. Acta Med. Port. 2022, 35, 270–278. [Google Scholar] [CrossRef]
- Groß, U.; Brzuszkiewicz, E.; Gunka, K.; Starke, J.; Riedel, T.; Bunk, B.; Spröer, C.; Wetzel, D.; Poehlein, A.; Chibani, C.; et al. Comparative genome and phenotypic analysis of three Clostridioides difficile strains isolated from a single patient provide insight into multiple infection of, C. difficile. BMC Genom. 2018, 19, 1. [Google Scholar] [CrossRef] [Green Version]
- Gerding, D.N.; Sambol, S.P.; Johnson, S. Non-toxigenic Clostridioides (Formerly Clostridium) difficile for Prevention of, C. difficile Infection: From Bench to Bedside Back to Bench and Back to Bedside. Front. Microbiol. 2018, 9, 1700. [Google Scholar] [CrossRef]
- Camorlinga, M.; Sanchez-Rojas, M.; Torres, J.; Romo-Castillo, M. Phenotypic Characterization of Non-toxigenic Clostridioides difficile Strains Isolated From Patients in Mexico. Front. Microbiol. 2019, 10, 84. [Google Scholar] [CrossRef]
- Dingle, K.E.; Griffiths, D.; Didelot, X.; Evans, J.; Vaughan, A.; Kachrimanidou, M.; Stoesser, N.; Jolley, K.A.; Golubchik, T.; Harding, R.M.; et al. Clinical Clostridium difficile: Clonality and pathogenicity locus diversity. PLoS ONE 2011, 6, e19993. [Google Scholar] [CrossRef]
- Stabler, R.A.; Gerding, D.N.; Songer, J.G.; Drudy, D.; Brazier, J.S.; Trinh, H.T.; Witney, A.A.; Hinds, J.; Wren, B.W. Comparative phylogenomics of Clostridium difficile reveals clade specificity and microevolution of hypervirulent strains. J. Bacteriol. 2006, 188, 7297–7305. [Google Scholar] [CrossRef] [Green Version]
- Neely, F.; Lambert, M.L.; Van Broeck, J.; Delmée, M. Clinical and laboratory features of the most common Clostridium difficile ribotypes isolated in Belgium. J. Hosp. Infect. 2017, 95, 394–399. [Google Scholar] [CrossRef]
- Muñoz, M.; Restrepo-Montoya, D.; Kumar, N.; Iraola, G.; Camargo, M.; Díaz-Arévalo, D.; Roa-Molina, N.S.; Tellez, M.A.; Herrera, G.; Ríos-Chaparro, D.I.; et al. Integrated genomic epidemiology and phenotypic profiling of Clostridium difficile across intra-hospital and community populations in Colombia. Sci. Rep. 2019, 9, 11293. [Google Scholar] [CrossRef] [Green Version]
- Drigo, I.; Mazzolini, E.; Bacchin, C.; Tonon, E.; Puiatti, C.; Bano, L.; Spigaglia, P.; Barbanti, F.; Agnoletti, F. Molecular characterization and antimicrobial susceptibility of Clostridium difficile isolated from rabbits raised for meat production. Vet. Microbiol. 2015, 181, 303–307. [Google Scholar] [CrossRef]
- Jacob, J.; Evers, S.; Bischoff, K.; Carlier, C.; Courvalin, P. Characterization of the sat4 gene encoding a streptothricin acetyltransferase in Campylobacter coli BE/G4. FEMS Microbiol. Lett. 1994, 120, 13–17. [Google Scholar] [CrossRef]
- Werner, G.; Hildebrandt, B.; Witte, W. Aminoglycoside-streptothricin resistance gene cluster aadE-sat4-aphA-3 disseminated among multiresistant isolates of Enterococcus faecium. Antimicrob. Agents Chemother. 2001, 45, 3267–3269. [Google Scholar] [CrossRef] [Green Version]
- Cafiso, V.; Lo Verde, F.; Zega, A.; Pigola, G.; Rostagno, R.; Borrè, S.; Stefani, S. Genomic Characterization of a New Biofilm-Forming and Adhesive ST398 Human-Adapted MSSA Lineage Causing Septic Knee Arthritis Following Surgical Reconstruction. Microorganisms 2021, 9, 305. [Google Scholar] [CrossRef]
- Suzuki, H.; Tomita, M.; Tsai, P.J.; Ko, W.C.; Hung, Y.P.; Huang, I.H.; Chen, J.W. Comparative genomic analysis of Clostridium difficile ribotype 027 strains including the newly sequenced strain NCKUH-21 isolated from a patient in Taiwan. Gut Pathog. 2017, 9, 70. [Google Scholar] [CrossRef] [Green Version]
- Knight, D.R.; Squire, M.M.; Collins, D.A.; Riley, T.V. Genome Analysis of Clostridium difficile PCR Ribotype 014 Lineage in Australian Pigs and Humans Reveals a Diverse Genetic Repertoire and Signatures of Long-Range Interspecies Transmission. Front. Microbiol. 2017, 7, 2138. [Google Scholar] [CrossRef] [Green Version]
- Lim, S.C.; Androga, G.O.; Knight, D.R.; Moono, P.; Foster, N.F.; Riley, T.V. Antimicrobial susceptibility of Clostridium difficile isolated from food and environmental sources in Western Australia. Int. J. Antimicrob. Agents 2018, 52, 411–415. [Google Scholar] [CrossRef] [PubMed]
- Spigaglia, P. Recent advances in the understanding of antibiotic resistance in Clostridium difficile infection. Ther. Adv. Infect. Dis. 2016, 3, 23–42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dridi, L.; Tankovic, J.; Petit, J. CdeA of Clostridium difficile, a new multidrug efflux transporter of the MATE family. Microb. Drug Resist. 2004, 10, 191–196. [Google Scholar] [CrossRef] [PubMed]
- O’Connor, J.R.; Galang, M.A.; Sambol, S.P.; Hecht, D.W.; Vedantam, G.; Gerding, D.N.; Johnson, S. Rifampin and rifaximin resistance in clinical isolates of Clostridium difficile. Antimicrob. Agents Chemother. 2008, 52, 2813–2817. [Google Scholar] [CrossRef] [Green Version]
- Toth, M.; Stewart, N.K.; Smith, C.; Vakulenko, S.B. Intrinsic Class D β-Lactamases of Clostridium difficile. mBio 2018, 6, e01803-18. [Google Scholar] [CrossRef] [Green Version]
- Stewart, N.K.; Smith, C.A.; Toth, M.; Stasyuk, A.; Vakulenko, S.B. The crystal structures of CDD-1, the intrinsic class D β-lactamase from the pathogenic Gram-positive bacterium Clostridioides difficile, and its complex with cefotaxime. J. Struct. Biol. 2019, 208, 107391. [Google Scholar] [CrossRef]
- Heir, E.; Sundheim, G.; Holck, A.L. The qacG gene on plasmid pST94 confers resistance to quaternary ammonium compounds in staphylococci isolated from the food industry. J. Appl. Microbiol. 1999, 86, 378–388. [Google Scholar] [CrossRef]
- Poole, K. Efflux pumps as antimicrobial resistance mechanisms. Ann. Med. 2007, 39, 162–176. [Google Scholar] [CrossRef]
- Wong, T.Z.; Zhang, M.; O’Donoghue, M.; Boost, M. Presence of antiseptic resistance genes in porcine methicillin-resistant Staphylococcus aureus. Vet. Microbiol. 2013, 162, 977–979. [Google Scholar] [CrossRef]
- Seier-Petersen, M.A.; Nielsen, L.N.; Ingmer, H.; Aarestrup, F.M.; Agersø, Y. Biocide Susceptibility of Staphylococcus aureus CC398 and CC30 Isolates from Pigs and Identification of the Biocide Resistance Genes, qacG and qacC. Microb. Drug Resist. 2015, 21, 527–536. [Google Scholar] [CrossRef]
- Ignak, S.; Nakipoglu, Y.; Gurler, B. Frequency of antiseptic resistance genes in clinical staphycocci and enterococci isolates in Turkey. Antimicrob. Resist. Infect. Control 2017, 30, 88. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.J.; Fu, L.; Huang, M.; Zhang, J.P.; Wu, Y.; Zhou, Y.S.; Zeng, J.; Wang, G.X. Frequency of antiseptic resistance genes and reduced susceptibility to biocides in carbapenem-resistant Acinetobacter baumannii. J. Med. Microbiol. 2017, 66, 13–17. [Google Scholar] [CrossRef]




| ID 1 | ST 2 | Clade | Toxin Genes 3 | |||
|---|---|---|---|---|---|---|
| tcdA | tcdB | cdtA | cdtB | |||
| 1 | 19 | 1 | + | + | − | − |
| 2 | 11 | 5 | + | + | + | + |
| 3 | 11 | 5 | + | + | + | + |
| 4 | 12 | 1 | + | + | − | − |
| 5 | 46 | 1 | + | + | − | − |
| 6 | 16 | 1 | + | + | − | − |
| 7 | 79 | 1 | − | − | − | − |
| 8 | 49 | 1 | + | + | − | − |
| Genotypic AMR Profile | ||||||
| ID | 23S rRNA Mutation (Erythromycin) | nimB Mutation 1 | 23S rRNA Mutation (Clindamycin) | cdeA | vanB | RpoB Mutation 2 |
| 1 | ||||||
| 2 | ||||||
| 3 | ||||||
| 4 | ||||||
| 5 | ||||||
| 6 | ||||||
| 7 | ||||||
| 8 | ||||||
| Erythromycin | Metronidazole | Clindamycin | Moxifloxacin | Vancomycin | Rifampicin | |
| Phenotypic AMR profile | ||||||
| Only in phenotypic AMR analysis | ||||||
| Only in genotypic AMR analysis | ||||||
| Found in both genotypic and phenotypic AMR analysis | ||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marcos, P.; Whyte, P.; Burgess, C.; Ekhlas, D.; Bolton, D. Detection and Genomic Characterisation of Clostridioides difficile from Spinach Fields. Pathogens 2022, 11, 1310. https://0-doi-org.brum.beds.ac.uk/10.3390/pathogens11111310
Marcos P, Whyte P, Burgess C, Ekhlas D, Bolton D. Detection and Genomic Characterisation of Clostridioides difficile from Spinach Fields. Pathogens. 2022; 11(11):1310. https://0-doi-org.brum.beds.ac.uk/10.3390/pathogens11111310
Chicago/Turabian StyleMarcos, Pilar, Paul Whyte, Catherine Burgess, Daniel Ekhlas, and Declan Bolton. 2022. "Detection and Genomic Characterisation of Clostridioides difficile from Spinach Fields" Pathogens 11, no. 11: 1310. https://0-doi-org.brum.beds.ac.uk/10.3390/pathogens11111310