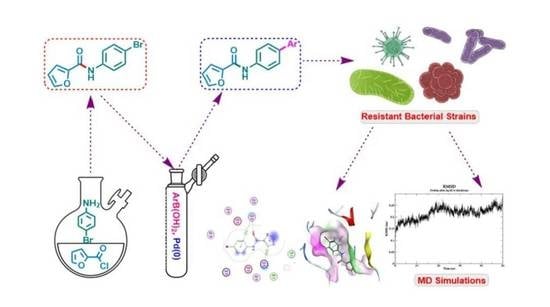
Synthesis of Functionalized N-(4-Bromophenyl)furan-2-carboxamides via Suzuki-Miyaura Cross-Coupling: Anti-Bacterial Activities against Clinically Isolated Drug Resistant A. baumannii, K. pneumoniae, E. cloacae and MRSA and Its Validation via a Computational Approach
Abstract
:1. Introduction
2. Results and Discussion
2.1. Chemistry
2.2. Antibacterial Activity
2.3. Molecular Docking Study
Molecular Docking Study of the NDM-1 A. baumannii/3, 5a-i Compounds
3. Materials and Methods
3.1. General Information
3.2. General Procedure for the Synthesis of Compounds
3.2.1. Synthesis of N-(4-Bromophenyl)furan-2-carboxamide (3)
3.2.2. Arylation of N-(4-Bromophenyl)furan-2-carboxamide (5a-i)
3.3. Characterization Data
3.3.1. N-(4-Bromophenyl)furan-2-carboxamide (3)
3.3.2. N-(3′-Chloro-4′-fluorobiphenyl-4-yl)furan-2-carboxamide (5a)
3.3.3. Methyl 4′-(Furan-2-carboxamido)biphenyl-4-carboxylate (5b)
3.3.4. N-(3′-Acetylbiphenyl-4-yl)furan-2-carboxamide (5c)
3.3.5. N-(4′-Methoxybiphenyl-4-yl)furan-2-carboxamide (5d)
3.3.6. N-(4-(Thiophen-3-yl)phenyl)furan-2-carboxamide (5e)
3.3.7. N-(4′-Chlorobiphenyl-4-yl)furan-2-carboxamide (5f)
3.3.8. N-(4-(Pyridin-3-yl)phenyl)furan-2-carboxamide (5g)
3.3.9. N-(3′,5′-Dichlorobiphenyl-4-yl)furan-2-carboxamide (5h)
3.3.10. N-(4′-Methylbiphenyl-4-yl)furan-2-carboxamide (5i)
3.4. Anti-Bacterial Activity
3.4.1. Identification of the Bacterial Strains
3.4.2. Antibiogram of the Isolates
3.4.3. Phenotypic Determination of MRSA
3.4.4. Phenotypic Detection of Carbapenemase Enzyme
3.4.5. Phenotypic Detection of Metallo-β-Lactamase (MBL) Enzyme
3.5. Antibacterial Activity of the Compound against XDR Pathogens
3.5.1. Agar Well Diffusion Method
3.5.2. Minimum Inhibitory Concertation of Different Compounds against XDR Pathogens
3.5.3. Determination of MBC
3.6. Docking Studies
3.7. Molecular Dynamics (MD) Simulation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Usman Qamar, M.; Lopes, B.S.; Hassan, B.; Khurshid, M.; Shafique, M.; Atif Nisar, M.; Mohsin, M.; Nawaz, Z.; Muzammil, S.; Aslam, B.; et al. The present danger of New Delhi metallo-β-lactamase: A threat to public health. Future Microbiol. 2020, 15, 1759–1778. [Google Scholar] [CrossRef] [PubMed]
- Aslam, B.; Wang, W.; Arshad, M.I.; Khurshid, M.; Muzammil, S.; Rasool, M.H.; Nisar, M.A.; Alvi, R.F.; Aslam, M.A.; Qamar, M.U.; et al. Antibiotic resistance: A rundown of a global crisis. Infect. Drug Resist. 2018, 11, 1645–1658. [Google Scholar] [CrossRef] [PubMed]
- Asokan, G.V.; Ramadhan, T.; Ahmed, E.; Sanad, H. WHO Global Priority Pathogens List: A Bibliometric Analysis of Medline-PubMed for Knowledge Mobilization to Infection Prevention and Control Practices in Bahrain. Oman Med. J. 2019, 34, 184–193. [Google Scholar] [CrossRef] [PubMed]
- Ejaz, H.; Younas, S.; Qamar, M.U.; Junaid, K.; Abdalla, A.E.; Abosalif, K.O.A.; Alameen, A.A.M.; Elamir, M.Y.M.; Ahmad, N.; Hamam, S.S.M.; et al. Molecular Epidemiology of Extensively Drug-Resistant mcr Encoded Colistin-Resistant Bacterial Strains Co-Expressing Multifarious β-Lactamases. Antibiotics 2021, 10, 467. [Google Scholar] [CrossRef] [PubMed]
- Bello-López, E.; del Carmen Rocha-Gracia, R.; Castro-Jaimes, S.; Cevallos, M.Á.; Vargas-Cruz, M.; Verdugo-Yocupicio, R.; Sáenz, Y.; Torres, C.; Gutiérrez-Cázarez, Z.; de la Paz Arenas, M.M. Antibiotic resistance mechanisms in Acinetobacter spp. strains isolated from patients in a paediatric hospital in Mexico. J. Glob. Antimicrob. Resist. 2020, 23, 120–129. [Google Scholar] [CrossRef]
- Liu, S.; Fang, R.; Zhang, Y.; Chen, L.; Huang, N.; Yu, K.; Zhou, C.; Cao, J.; Zhou, T. Characterization of resistance mechanisms of Enterobacter cloacae Complex co-resistant to carbapenem and colistin. BMC Microbiol. 2021, 21, 208. [Google Scholar] [CrossRef]
- Eger, E.; Schwabe, M.; Schulig, L.; Hübner, N.-O.; Bohnert, J.A.; Bornscheuer, U.T.; Heiden, S.E.; Müller, J.U.; Adnan, F.; Becker, K. Extensively Drug-Resistant Klebsiella pneumoniae Counteracts Fitness and Virulence Costs That Accompanied Ceftazidime-Avibactam Resistance Acquisition. Microbiol. Spectr. 2022, 10, e00148-22. [Google Scholar] [CrossRef]
- Liu, W.-T.; Chen, E.-Z.; Yang, L.; Peng, C.; Wang, Q.; Xu, Z.; Chen, D.-Q. Emerging resistance mechanisms for 4 types of common anti-MRSA antibiotics in Staphylococcus aureus: A comprehensive review. Microb. Pathog. 2021, 156, 104915. [Google Scholar] [CrossRef]
- Gabriel, G.J.; Som, A.; Madkour, A.E.; Eren, T.; Tew, G.N. Infectious disease: Connecting innate immunity to biocidal polymers. Mater. Sci. Eng. R Rep. 2007, 57, 28–64. [Google Scholar] [CrossRef]
- Phillips, D.J.; Harrison, J.; Richards, S.-J.; Mitchell, D.E.; Tichauer, E.; Hubbard, A.T.; Guy, C.; Hands-Portman, I.; Fullam, E.; Gibson, M.I. Evaluation of the antimicrobial activity of cationic polymers against mycobacteria: Toward antitubercular macromolecules. Biomacromolecules 2017, 18, 1592–1599. [Google Scholar] [CrossRef]
- Tuchilus, C.G.; Nichifor, M.; Mocanu, G.; Stanciu, M.C. Antimicrobial activity of chemically modified dextran derivatives. Carbohydr. Polym. 2017, 161, 181–186. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; Suzuki, M.; Xie, L.; Morris-Natschke, S.L.; Lee, K.H. Recent progress in the development of coumarin derivatives as potent anti-HIV agents. Med. Res. Rev. 2003, 23, 322–345. [Google Scholar] [CrossRef] [PubMed]
- Buzzini, P.; Turchetti, B.; Ieri, F.; Goretti, M.; Branda, E.; Mulinacci, N.; Romani, A. Catechins and proanthocyanidins: Naturally occurring O-heterocycles with antimicrobial activity. In Bioactive Heterocycles IV; Springer: Berlin/Heidelberg, Germany, 2007; pp. 239–263. [Google Scholar]
- Donnelly, D.; Boland, G. Isoflavonoids and neoflavonoids: Naturally occurring O-heterocycles. Nat. Prod. Rep. 1995, 12, 321–338. [Google Scholar] [CrossRef]
- Kumawat, M.K. Thiazole containing heterocycles with antimalarial activity. Curr. Drug Discov. Technol. 2018, 15, 196–200. [Google Scholar] [CrossRef] [PubMed]
- Iriepa, I.; Madrid, A.; Gálvez, E.; Bellanato, J. Synthesis, structural and conformational study of some amides derived from N-methylpiperazine. J. Mol. Struct. 2006, 787, 8–13. [Google Scholar] [CrossRef]
- Ouyang, X.; Armstrong, R.W.; Murphy, M.M. A novel cleavage technique to generate small molecule compounds and libraries via a two-resin system. J. Org. Chem. 1998, 63, 1027–1032. [Google Scholar] [CrossRef]
- Gudasi, K.B.; Vadavi, R.S.; Shenoy, R.V.; Patil, M.S.; Patil, S.A. Ambidentate behavior of pentadentate N, N′-bis (2-hydroxyphenyl)-pyridine-2, 6-dicarboxamide towards transition metal (II) salts. Transit. Met. Chem. 2005, 30, 569–574. [Google Scholar] [CrossRef]
- Auzeil, N.; Largeron, M.; Fleury, M.-B. Acid-catalysed N–alkyl heterolysis of tertiary pyridinecarboxamides and benzamides under mild conditions. J. Chem. Soc. Perkin Trans. 1999, 2, 1703–1710. [Google Scholar] [CrossRef]
- Antoszczak, M.; Maj, E.; Stefańska, J.; Wietrzyk, J.; Janczak, J.; Brzezinski, B.; Huczyński, A. Synthesis, antiproliferative and antibacterial activity of new amides of salinomycin. Bioorg. Med. Chem. Lett. 2014, 24, 1724–1729. [Google Scholar] [CrossRef]
- Antoszczak, M.; Maj, E.; Napiórkowska, A.; Stefańska, J.; Augustynowicz-Kopeć, E.; Wietrzyk, J.; Janczak, J.; Brzezinski, B.; Huczyński, A. Synthesis, anticancer and antibacterial activity of salinomycin N-benzyl amides. Molecules 2014, 19, 19435–19459. [Google Scholar] [CrossRef]
- Singh, M.; Singh, S.K.; Gangwar, M.; Nath, G.; Singh, S.K. Design, synthesis and mode of action of some benzothiazole derivatives bearing an amide moiety as antibacterial agents. RSC Adv. 2014, 4, 19013–19023. [Google Scholar] [CrossRef]
- Blangetti, M.; Rosso, H.; Prandi, C.; Deagostino, A.; Venturello, P. Suzuki-Miyaura cross-coupling in acylation reactions, scope and recent developments. Molecules 2013, 18, 1188–1213. [Google Scholar] [CrossRef] [PubMed]
- Han, F.-S. Transition-metal-catalyzed Suzuki–Miyaura cross-coupling reactions: A remarkable advance from palladium to nickel catalysts. Chem. Soc. Rev. 2013, 42, 5270–5298. [Google Scholar] [CrossRef] [PubMed]
- Billingsley, K.; Buchwald, S.L. Highly efficient monophosphine-based catalyst for the palladium-catalyzed suzuki− miyaura reaction of heteroaryl halides and heteroaryl boronic acids and esters. J. Am. Chem. Soc. 2007, 129, 3358–3366. [Google Scholar] [CrossRef]
- Malik, A.; Rasool, N.; Kanwal, I.; Hashmi, M.A.; Zahoor, A.F.; Ahmad, G.; Altaf, A.A.; Shah, S.A.A.; Sultan, S.; Zakaria, Z.A. Suzuki–Miyaura Reactions of (4-bromophenyl)-4, 6-dichloropyrimidine through Commercially Available Palladium Catalyst: Synthesis, Optimization and Their Structural Aspects Identification through Computational Studies. Processes 2020, 8, 1342. [Google Scholar] [CrossRef]
- Imran, H.M.; Rasool, N.; Kanwal, I.; Hashmi, M.A.; Altaf, A.A.; Ahmed, G.; Malik, A.; Kausar, S.; Khan, S.U.-D.; Ahmad, A. Synthesis of halogenated [1, 1′-biphenyl]-4-yl benzoate and [1, 1′: 3′, 1″-terphenyl]-4′-yl benzoate by palladium catalyzed cascade C–C coupling and structural analysis through computational approach. J. Mol. Struct. 2020, 1222, 128839. [Google Scholar] [CrossRef]
- Ning, N.-Z.; Liu, X.; Chen, F.; Zhou, P.; Hu, L.; Huang, J.; Li, Z.; Huang, J.; Li, T.; Wang, H. Embelin restores carbapenem efficacy against NDM-1-positive pathogens. Front. Microbiol. 2018, 9, 71. [Google Scholar] [CrossRef]
- Gonec, T.; Kos, J.; Nevin, E.; Govender, R.; Pesko, M.; Tengler, J.; Kushkevych, I.; Stastna, V.; Oravec, M.; Kollar, P. Preparation and biological properties of ring-substituted naphthalene-1-carboxanilides. Molecules 2014, 19, 10386–10409. [Google Scholar] [CrossRef]
- Kanwal, I.; Rasool, N.; Zaidi, S.H.M.; Zakaria, Z.A.; Bilal, M.; Hashmi, M.A.; Mubarik, A.; Ahmad, G.; Shah, S.A.A. Synthesis of Functionalized Thiophene Based Pyrazole Amides via Various Catalytic Approaches: Structural Features through Computational Applications and Nonlinear Optical Properties. Molecules 2022, 27, 360. [Google Scholar] [CrossRef]
- Loganathan, A.; Manohar, P.; Eniyan, K.; Jayaraj, R.; Nachimuthu, R. Evaluation of various phenotypic methods with genotypic screening for detection of methicillin-resistant. Asian Biomed. 2019, 13, 225–233. [Google Scholar] [CrossRef]
- Qamar, M.U.; Walsh, T.R.; Toleman, M.A.; Tyrrell, J.M.; Saleem, S.; Aboklaish, A.; Jahan, S. Dissemination of genetically diverse NDM-1, -5, -7 producing-Gram-negative pathogens isolated from pediatric patients in Pakistan. Future Microbiol. 2019, 14, 691–704. [Google Scholar] [CrossRef] [PubMed]
- Qamar, M.U.; Saleem, S.; Toleman, M.A.; Saqalein, M.; Waseem, M.; Nisar, M.A.; Khurshid, M.; Taj, Z.; Jahan, S. In vitro and in vivo activity of Manuka honey against NDM-1-producing Klebsiella pneumoniae ST11. Future Microbiol. 2018, 13, 13–26. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, G.; Rasool, N.; Qamar, M.U.; Alam, M.M.; Kosar, N.; Mahmood, T.; Imran, M. Facile synthesis of 4-aryl-N-(5-methyl-1H-pyrazol-3-yl) benzamides via Suzuki Miyaura reaction: Antibacterial activity against clinically isolated NDM-1-positive bacteria and their Docking Studies. Arab. J. Chem. 2021, 14, 103270. [Google Scholar] [CrossRef]
- Verdonk, M.L.; Cole, J.C.; Hartshorn, M.J.; Murray, C.W.; Taylor, R.D. Improved protein–ligand docking using GOLD. Proteins Struct. Funct. Bioinform. 2003, 52, 609–623. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The protein data bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Al-Karmalawy, A.A.; Dahab, M.A.; Metwaly, A.M.; Elhady, S.S.; Elkaeed, E.B.; Eissa, I.H.; Darwish, K.M. Molecular docking and dynamics simulation revealed the potential inhibitory activity of ACEIs against SARS-CoV-2 targeting the hACE2 receptor. Front. Chem. 2021, 9, 661230. [Google Scholar] [CrossRef]
- Van Der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J. GROMACS: Fast, flexible, and free. J. Comput. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef]
- Assadollahi, V.; Rashidieh, B.; Alasvand, M.; Abdolahi, A.; Lopez, J.A. Interaction and molecular dynamics simulation study of Osimertinib (AstraZeneca 9291) anticancer drug with the EGFR kinase domain in native protein and mutated L844V and C797S. J. Cell. Biochem. 2019, 120, 13046–13055. [Google Scholar] [CrossRef]







| Comp. No. | Bacterial Strains | Zone (mm) (50 mg) | Zone (mm) (40 mg) | Zone (mm) (30 mg) | Zone (mm) (20 mg) | Zone (mm) (10 mg) | Zone (mm) DMSO | Zone (mm) Meropenem |
|---|---|---|---|---|---|---|---|---|
| 3 | CRAB | 18 ± 1 | 11 ± 2 | 7 ± 2 | 6 ± 2 | 4 ± 0 | 0 ± 0 | 4 |
| CREC | 10 ± 2 | 8 ± 3 | 7 ± 3 | 5 ± 2 | 4 ± 0 | 0 ± 0 | 4 | |
| CRKP | 8 ± 1 | 7 ± 2 | 6 ± 2 | 5 ± 2 | 4 ± 0 | 0 ± 0 | 4 | |
| MRSA | 11 ± 2 | 9 ± 1 | 7 ± 2 | 6 ± 2 | 5 ± 0 | 0 ± 0 | 4 | |
| 5c | CRAB | 6 ± 3 | 4 ± 2 | 4 ± 2 | 4 ± 3 | 4 ± 0 | 0 ± 0 | 4 |
| CREC | 5 ± 2 | 4 ± 1 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 4 | |
| CRKP | 4 ± 1 | 4 ± 2 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 4 | |
| MRSA | 5 ± 1 | 4 ± 2 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 4 | |
| 5d | CRAB | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 4 |
| CREC | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 4 | |
| CRKP | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 4 | |
| MRSA | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 4 | |
| 5g | CRAB | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 4 |
| CREC | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 4 | |
| CRKP | 4 ± 2 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 4 | |
| MRSA | 4 ± 3 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 4 | |
| 5h | CRAB | 5 ± 2 | 4 ± 2 | 4 ± 2 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 4 |
| CREC | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 4 | |
| CRKP | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 4 | |
| MRSA | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 4 | |
| 5i | CRAB | 4 ± 2 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 4 |
| CREC | 5 ± 1 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 4 | |
| CRKP | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 4 | |
| MRSA | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 4 |
| Pathogens | MIC | MBC |
|---|---|---|
| CRAB | 6.25 | 12.5 |
| CREC | 6.25 | 12.5 |
| CRKP | 6.25 | 12.5 |
| MRSA | 12.5 | 25 |
| Antibiotics | MIC Breakpoints (µg/mL) | A. baumannii | K. pneumoniae | E. cloacae | S. aureus |
|---|---|---|---|---|---|
| P | ≤0.12–≥0.25 | NA | NA | NA | ≥16 |
| E | ≤0.5–≥8 | NA | NA | NA | ≥32 |
| DA | ≤0.5–≥4 | NA | NA | NA | ≥16 |
| SAM | ≤8/4–≥32/16 | ≥64/128 | ≥64/32 | ≥64/128 | NA |
| PIP | ≤16–≥128 | ≥256 | ≥256 | ≥256 | NA |
| FOX | ≤4–≥8 | NA | NA | NA | ≥32 |
| CRO | ≤8–≥64 | ≥256 | ≥256 | ≥128 | ≥128 |
| FEP | ≤8–≥32 | ≥256 | ≥128 | ≥128 | ≥64 |
| ATM | ≤4–≥16 | ≥64 | ≥64 | ≥64 | ≥64 |
| MEM | ≤2–≥8 | ≥32 | ≥32 | ≥32 | ≥8 |
| LEV | ≤1–≥4 | ≥16 | ≥16 | ≥16 | 0.5 |
| MXF | ≤0.5–≥2 | ≥16 | ≥16 | ≥16 | NA |
| MNO | ≤4–≥16 | ≥64 | ≥64 | ≥32 | NA |
| TE | ≤4–≥16 | ≥32 | ≥32 | ≥32 | NA |
| C | ≤8–≥32 | ≥64 | ≥128 | ≥64 | NA |
| SXT | ≤2/38–≥4/76 | ≥8/304 | ≥8/304 | ≥8/304 | ≥8/152 |
| VA | ≤2–≥16 | NA | NA | NA | 1 |
| LZD | ≤4–≥8 | NA | NA | NA | 2 |
| CS | ≥4 | 0.5 | 0.5 | 0.5 | NA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Siddiqa, A.; Zubair, M.; Bilal, M.; Rasool, N.; Qamar, M.U.; Khalid, A.; Ahmad, G.; Imran, M.; Mahmood, S.; Ashraf, G.A. Synthesis of Functionalized N-(4-Bromophenyl)furan-2-carboxamides via Suzuki-Miyaura Cross-Coupling: Anti-Bacterial Activities against Clinically Isolated Drug Resistant A. baumannii, K. pneumoniae, E. cloacae and MRSA and Its Validation via a Computational Approach. Pharmaceuticals 2022, 15, 841. https://0-doi-org.brum.beds.ac.uk/10.3390/ph15070841
Siddiqa A, Zubair M, Bilal M, Rasool N, Qamar MU, Khalid A, Ahmad G, Imran M, Mahmood S, Ashraf GA. Synthesis of Functionalized N-(4-Bromophenyl)furan-2-carboxamides via Suzuki-Miyaura Cross-Coupling: Anti-Bacterial Activities against Clinically Isolated Drug Resistant A. baumannii, K. pneumoniae, E. cloacae and MRSA and Its Validation via a Computational Approach. Pharmaceuticals. 2022; 15(7):841. https://0-doi-org.brum.beds.ac.uk/10.3390/ph15070841
Chicago/Turabian StyleSiddiqa, Ayesha, Muhammad Zubair, Muhammad Bilal, Nasir Rasool, Muhammad Usman Qamar, Aqsa Khalid, Gulraiz Ahmad, Muhammad Imran, Sajid Mahmood, and Ghulam Abbas Ashraf. 2022. "Synthesis of Functionalized N-(4-Bromophenyl)furan-2-carboxamides via Suzuki-Miyaura Cross-Coupling: Anti-Bacterial Activities against Clinically Isolated Drug Resistant A. baumannii, K. pneumoniae, E. cloacae and MRSA and Its Validation via a Computational Approach" Pharmaceuticals 15, no. 7: 841. https://0-doi-org.brum.beds.ac.uk/10.3390/ph15070841