Processing Chain for Estimation of Tree Diameter from GNSS-IMU-Based Mobile Laser Scanning Data
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Site
2.2. Mobile Mapping System
2.3. Mobile Laser Scanning Data
2.4. Reference Data
2.5. Data Processing
2.5.1. Pre-Processing of Mobile Mapping System Data
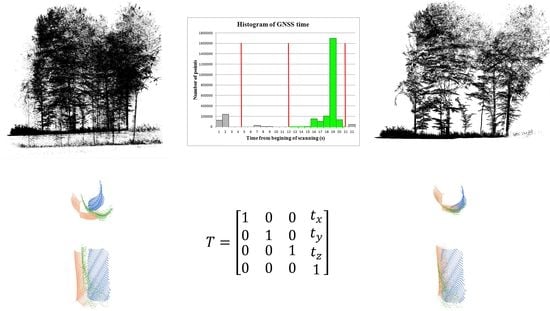
2.5.2. GNSS Time-Based Point Clustering
2.5.3. Extraction of Tree Stems
2.5.4. Matching of Point Clusters from Individual Tree Stems
2.5.5. Registration of Point Clusters from Individual Tree Stems
I. Finding the Overlap
II. Selection
III. Matching
IV. Rejection
- Maximum roughness of fitted planes was higher than 0.15 and 0.4 for noise-free and noisy point clusters, respectively.
- Maximum angle deviations between the normals of focal neighborhoods was higher than 15° or 40° for noise-free and noisy point clusters, respectively.
V. Weighting
VI. Minimization
VII. Transformation
2.5.6. Estimation of Diameter at Breast Height
2.5.7. Estimation of MLS Coverage of Tree Stems
3. Results
3.1. GNSS Time-Based Clustering
3.2. Matching and Registration of Point Clusters from Individual Tree Stems
3.3. Estimation of Diameter at Breast Height
3.4. MLS Coverage of Tree Stems
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Optech Inc. Mobile Mapping System Lynx. 2007. Available online: https://www.teledyneoptech.com (accessed on 12 March 2019).
- MDL Laser Systems 2010. Mobile Mapping System Dynascan. Available online: https://www.renishaw.com (accessed on 12 March 2019).
- RIEGL Laser Measurement Systems GmbH 2010. Mobile Mapping System Riegl VMX-250. Available online: http://www.riegl.com (accessed on 12 March 2019).
- Topcon Corporation 2010. Mobile Mapping System IP-S2. Available online: https://www.topcon.co.jp/ (accessed on 12 March 2019).
- SITECO Inf. Mobile mapping system Roadscanner. 2012. Available online: www.sitecoinf.it/it/ (accessed on 12 March 2019).
- Puente, I.; González-Jorge, H.; Martínez-Sánchez, J.; Arias, P. Review of mobile mapping and surveying technologies. Meas. J. Int. Meas. Confed. 2013, 46, 2127–2145. [Google Scholar] [CrossRef]
- Liang, X.; Hyyppa, J.; Kukko, A.; Kaartinen, H.; Jaakkola, A.; Yu, X. The use of a mobile laser scanning system for mapping large forest plots. IEEE Geosci. Remote Sens. Lett. 2014, 11, 1504–1508. [Google Scholar] [CrossRef]
- Liang, X.; Kukko, A.; Kaartinen, H.; Hyyppä, J.; Yu, X.; Jaakkola, A.; Wang, Y. Possibilities of a personal laser scanning system for forest mapping and ecosystem services. Sensors 2014, 14, 1228–1248. [Google Scholar] [CrossRef] [PubMed]
- Rönnholm, P.; Liang, X.; Kukko, A.; Jaakkola, A.; Hyyppä, J. Quality analysis and correction of mobile backpack laser scanning data. ISPRS Ann. Photogramm. Remote Sens. Spat. Inf. Sci. 2016, 3, 41–47. [Google Scholar] [CrossRef]
- Kukko, A.; Kaijaluoto, R.; Kaartinen, H.; Lehtola, V.V.; Jaakkola, A.; Hyyppä, J. Graph SLAM correction for single scanner MLS forest data under boreal forest canopy. ISPRS J. Photogramm. Remote Sens. 2017, 132, 199–209. [Google Scholar] [CrossRef]
- Pierzchała, M.; Giguère, P.; Astrup, R. Mapping forests using an unmanned ground vehicle with 3D LiDAR and graph-SLAM. Comput. Electron. Agric. 2018, 145, 217–225. [Google Scholar] [CrossRef]
- Ryding, J.; Williams, E.; Smith, M.J.; Eichhorn, M.P. Assessing handheld mobile laser scanners for forest surveys. Remote Sens. 2015, 7, 1095–1111. [Google Scholar] [CrossRef]
- Tang, J.; Chen, Y.; Kukko, A.; Kaartinen, H.; Jaakkola, A.; Khoramshahi, E.; Hakala, T.; Hyyppä, J.; Holopainen, M.; Hyyppä, H. SLAM-aided stem mapping for forest inventory with small-footprint mobile LiDAR. Forests 2015, 6, 4588–4606. [Google Scholar] [CrossRef]
- Bauwens, S.; Bartholomeus, H.; Calders, K.; Lejeune, P. Forest inventory with terrestrial LiDAR: A comparison of static and hand-held mobile laser scanning. Forests 2016, 7, 127. [Google Scholar] [CrossRef]
- Forsman, M.; Holmgren, J.; Olofsson, K. Tree stem diameter estimation from mobile laser scanning using line-wise intensity-based clustering. Forests 2016, 7, 206. [Google Scholar] [CrossRef]
- Cabo, C.; Del Pozo, S.; Rodríguez-Gonzálvez, P.; Ordóñez, C.; González-Aguilera, D. Comparing terrestrial laser scanning (TLS) and wearable laser scanning (WLS) for individual tree modeling at plot level. Remote Sens. 2018, 10, 540. [Google Scholar] [CrossRef]
- Puttonen, E.; Jaakkola, A.; Litkey, P.; Hyyppä, J. Tree classification with fused mobile laser scanning and hyperspectral data. Sensors 2011, 11, 5158–5182. [Google Scholar] [CrossRef]
- Holopainen, M.; Kankare, V.; Vastaranta, M.; Liang, X.; Lin, Y.; Vaaja, M.; Yu, X.; Hyyppä, J.; Hyyppä, H.; Kaartinen, H. Tree mapping using airborne, terrestrial and mobile laser scanning–A case study in a heterogeneous urban forest. Urban For. Urban Green. 2013, 12, 546–553. [Google Scholar] [CrossRef]
- Wu, B.; Yu, B.; Yue, W.; Shu, S.; Tan, W.; Hu, C.; Huang, Y.; Wu, J.; Liu, H. A voxel-based method for automated identification and morphological parameters estimation of individual street trees from mobile laser scanning data. Remote Sens. 2013, 5, 584–611. [Google Scholar] [CrossRef]
- Čerňava, J.; Tuček, J.; Koreň, M.; Mokroš, M. Estimation of diameter at breast height from mobile laser scanning data collected under a heavy forest canopy. J. For. Sci. 2017, 63, 433–441. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Medioni, G. Object modelling by registration of multiple range images. Image Vis. Comput. 1992, 10, 145–155. [Google Scholar] [CrossRef]
- Besl, P.J.; McKay, N.D. A Method for Registration of 3-D Shapes. Trans. Pattern Anal. Mach. Intell. 1991, 14, 239–256. [Google Scholar] [CrossRef]
- MathWorks, M. SIMULINK for Technical Computing. 2015. Available online: https://www.mathworks.com/ (accessed on 12 March 2019).
- CloudCompare. GPL Software.; (version 2.6.2). Available online: http://www.cloudcompare.org/ (accessed on 12 March 2019).
- Glira, P.; Pfeifer, N.; Briese, C.; Ressl, C. A Correspondence Framework for ALS Strip Adjustments based on Variants of the ICP Algorithm. Photogramm. Fernerkundung Geoinf. 2015, 2015, 275–289. [Google Scholar] [CrossRef]
- Glira, P.; Pfeifer, N.; Mandlburger, G. Rigorous Strip adjustment of UAV-based laserscanning data including time-dependent correction of trajectory errors. Photogramm. Eng. Remote Sens. 2016, 82, 945–954. [Google Scholar] [CrossRef]
- Liang, X.; Kukko, A.; Hyyppä, J.; Lehtomäki, M.; Pyörälä, J.; Yu, X.; Kaartinen, H.; Jaakkola, A.; Wang, Y. In-situ measurements from mobile platforms: An emerging approach to address the old challenges associated with forest inventories. ISPRS J. Photogramm. Remote Sens. 2018, 143, 97–107. [Google Scholar] [CrossRef]
- Koreň, M.; Mokroš, M.; Bucha, T. Accuracy of tree diameter estimation from terrestrial laser scanning by circle-fitting methods. Int. J. Appl. Earth Obs. Geoinf. 2017, 63, 122–128. [Google Scholar] [CrossRef]
- Mandlburger, G.; Otepka, J.; Karel, W.; Wagner, W.; Pfeifer, N. Orientation and processing of airborne laser scanning data (OPALS)—Concept and first results of a comprehensive ALS software. IAPRS 2009, XXXVIII, 55–60. [Google Scholar]
- Otepka, J.; Mandlburger, G.; Karel, W. The OPALS data manager—Efficient data management for processing large airborne laser scanning projects. ISPRS Ann. Photogramm. Remote Sens. Spat. Inf. Sci. 2012, 1, 153–159. [Google Scholar] [CrossRef]
- Pfeifer, N.; Mandlburger, G.; Otepka, J.; Karel, W. OPALS—A framework for Airborne Laser Scanning data analysis. Comput. Environ. Urban Syst. 2014, 45, 125–136. [Google Scholar] [CrossRef]
- Simanov, V.; Macků, J.; Popelka, J. Proposal of a new terrain classification and technological typification. Lesnictví 1993, 39, 422–428. [Google Scholar]
- Boavida, J.; Oliveira, A.; Santos, B. Precise long tunnel survey using the Riegl VMX-250 mobile laser scanning system. In Proceedings of the 2012 RIEGL International Airborne and Mobile User Conference, Orlando, FL, USA, 27 February–1 March 2012; Volume 27. [Google Scholar]
- Isenburg, M. LAStools—Efficient LiDAR Processing Software, (version 160429, Academic); rapidlasso GmbH: Gilching, Germany, 2016. [Google Scholar]
- Rusinkiewicz, S.; Levoy, M. Efficient variants of the ICP algorithm. In Proceedings of the Third International Conference on 3-D Digital Imaging and Modeling, Quebec City, QC, Canada, 28 May–1 June 2001; pp. 145–152. [Google Scholar]
- Thies, M.; Spiecker, H.; Str, T. Evaluation and future prospects of terrestrial laser scanning for standardized laser scanning for standardized forest inventories. Int. Arch. Photogramm. Remote Sens. 2004, 36, 192–197. [Google Scholar]
- Wang, D.; Hollaus, M.; Puttonen, E.; Pfeifer, N. Automatic and Self-Adaptive Stem Reconstruction in Landslide-Affected Forests. Remote Sens. 2016, 8, 974. [Google Scholar] [CrossRef]
- Liang, X.; Litkey, P.; Hyyppa, J.; Kaartinen, H.; Vastaranta, M.; Holopainen, M. Automatic stem mapping using single-scan terrestrial laser scanning. IEEE Trans. Geosci. Remote Sens. 2012, 50, 661–670. [Google Scholar] [CrossRef]

















| MMS Physical Data | Main Dimension (L × W × H) (mm) | Weight (kg) |
|---|---|---|
| VMX-250-MH Measuring head | 737 × 456 × 485 | 43 |
| VMX-250-CU Control Unit | 560 × 455 × 265 | 26 |
| VMX-250-CS6 Camera System | 607 × 1038 × 208 | 19 |
| GNSS-IMU Performance | |
| Parameter | Value |
| Position (absolute) (mm) | 20–50 |
| Position (relative) (mm) | 10 |
| Roll and Pitch (°) | 0.005 |
| Heading (°) | 0.015 |
| Mobile Laser Scanner VQ-250 | |
| Parameter | Value |
| Laser Measuring Principle | Time of flight |
| Horizontal Field of View (°) | 360 |
| Vertical Field of View (°) | 360 |
| Minimum Measurement Range (m) | 1.5 |
| Maximum Measurement Range (m) | 200 |
| Accuracy (mm) | 10 |
| Precision (mm) | 5 |
| Laser Pulse Repetition Rate (kHz) | 300 |
| Max. Effective Measurement Rate (pnts/s) | 300000 |
| Laser Wavelength | Near-infrared |
| Laser Beam Divergence (mrad) | 0.35 |
| Laser Beam Footprint (mm/50 m) | 18 |
| Angle Measurement Resolution (°) | 0.001 |
| Processing Step | Matching | Overlap Calculation | Registration | ||||
|---|---|---|---|---|---|---|---|
| Parameters | dist. | dist. + id | dist. + id + ∆d | ||||
| Sample size | 779 | 779 | 779 | 531 | 430 | ||
| Accuracy | Absolute | 627 | 674 | 660 | 514 | 407 | |
| Relative (%) | 80.49 | 86.52 | 84.72 | 96.8 | 94.65 | ||
| Errors | Commission | Number | 152 | 95 | 24 | 3 | 23 |
| Percentage (%) | 19.51 | 12.2 | 3.08 | 0.56 | 2.9 | ||
| Omission | Number | 0 | 10 | 95 | 14 | 0 | |
| Percentage (%) | 0 | 1.28 | 12.2 | 2.64 | 0 | ||
| N | p-Value | emin (cm) | emax (cm) | RMSE (cm) | RMSE% (%) | ||
|---|---|---|---|---|---|---|---|
| 154 | 45.02 | 0.0091 | −7.78 | 9.16 | 0.63 | 3.06 | 6.71 |
| MLS Coverage (%) | N | RMSE (cm) | %RMSE (%) | ||
|---|---|---|---|---|---|
| 20–30 | 5 | 58.30 | −2.39 | 3.53 | 6.06 |
| 30–40 | 7 | 54.08 | 1.44 | 6.74 | 12.46 |
| 40–50 | 38 | 44.88 | 1.22 | 3.11 | 6.92 |
| 50–60 | 45 | 42.68 | 0.73 | 3.13 | 7.33 |
| 60–70 | 44 | 46.51 | 0.84 | 2.05 | 4.41 |
| 70–80 | 11 | 38.63 | −1.14 | 2.17 | 5.61 |
| 80+ | 4 | 41.31 | −1.34 | 2.19 | 5.31 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Čerňava, J.; Mokroš, M.; Tuček, J.; Antal, M.; Slatkovská, Z. Processing Chain for Estimation of Tree Diameter from GNSS-IMU-Based Mobile Laser Scanning Data. Remote Sens. 2019, 11, 615. https://0-doi-org.brum.beds.ac.uk/10.3390/rs11060615
Čerňava J, Mokroš M, Tuček J, Antal M, Slatkovská Z. Processing Chain for Estimation of Tree Diameter from GNSS-IMU-Based Mobile Laser Scanning Data. Remote Sensing. 2019; 11(6):615. https://0-doi-org.brum.beds.ac.uk/10.3390/rs11060615
Chicago/Turabian StyleČerňava, Juraj, Martin Mokroš, Ján Tuček, Michal Antal, and Zuzana Slatkovská. 2019. "Processing Chain for Estimation of Tree Diameter from GNSS-IMU-Based Mobile Laser Scanning Data" Remote Sensing 11, no. 6: 615. https://0-doi-org.brum.beds.ac.uk/10.3390/rs11060615